Identification of an Intramolecular Switch That Controls the Interaction of Helicase nsp10 with Membrane-Associated nsp12 of Porcine Reproductive and Respiratory Syndrome Virus
- PMID: 34076477
- PMCID: PMC8354221
- DOI: 10.1128/JVI.00518-21
Identification of an Intramolecular Switch That Controls the Interaction of Helicase nsp10 with Membrane-Associated nsp12 of Porcine Reproductive and Respiratory Syndrome Virus
Abstract
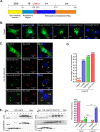
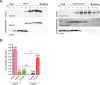
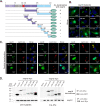
A critical step in replication of positive-stranded RNA viruses is the assembly of replication and transcription complexes (RTC). We have recently mapped the nonstructural protein (nsp) interaction network of porcine reproductive and respiratory syndrome virus (PRRSV) and provided evidence by truncation mutagenesis that the recruitment of viral core replicase enzymes (nsp9 and nsp10) to membrane proteins (nsp2, nsp3, nsp5, and nsp12) is subject to regulation. Here, we went further to discover an intramolecular switch within the helicase nsp10 that controls its interaction with the membrane-associated protein nsp12. Deletion of nsp10 linker region amino acids 124 to 133, connecting domain 1B to 1A, led to complete relocalization and colocalization in the cells coexpressing nsp12. Moreover, single-amino-acid substitutions (e.g., nsp10 E131A and I132A) were sufficient to enable the nsp10-nsp12 interaction. Further proof came from membrane floatation assays that revealed a clear movement of nsp10 mutants, but not wild-type nsp10, toward the top of sucrose gradients in the presence of nsp12. Interestingly, the same mutations were not able to activate the nsp10-nsp2/3 interaction, suggesting a differential requirement for conformation. Reverse genetics analysis showed that PRRSV mutants carrying the single substitutions were not viable and were defective in subgenomic RNA (sgRNA) accumulation. Together, our results provide strong evidence for a regulated interaction between nsp10 and nsp12 and suggest an essential role for an orchestrated RTC assembly in sgRNA synthesis. IMPORTANCE Assembly of replication and transcription complexes (RTC) is a limiting step for viral RNA synthesis. The PRRSV RTC macromolecular complexes are comprised of mainly viral nonstructural replicase proteins (nsps), but how they come together remains elusive. We previously showed that viral helicase nsp10 interacts nsp12 in a regulated manner by truncation mutagenesis. Here, we revealed that the interaction is controlled by single residues within the domain linker region of nsp10. Moreover, the activation mutations lead to defects in viral sgRNA synthesis. Our results provide important insight into the mechanisms of PRRSV RTC assembly and regulation of viral sgRNA synthesis.
Keywords: assembly; nonstructural protein; porcine reproductive and respiratory syndrome virus; regulated interaction; replication and transcription complexes; subgenomic RNA synthesis.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

