NAP1L1 and NAP1L4 Binding to Hypervariable Domain of Chikungunya Virus nsP3 Protein Is Bivalent and Requires Phosphorylation
- PMID: 34076483
- PMCID: PMC8312869
- DOI: 10.1128/JVI.00836-21
NAP1L1 and NAP1L4 Binding to Hypervariable Domain of Chikungunya Virus nsP3 Protein Is Bivalent and Requires Phosphorylation
Abstract
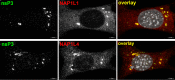
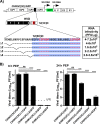
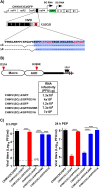
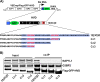
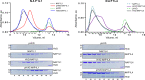
Chikungunya virus (CHIKV) is one of the most pathogenic members of the Alphavirus genus in the Togaviridae family. Within the last 2 decades, CHIKV has expanded its presence to both hemispheres and is currently circulating in both Old and New Worlds. Despite the severity and persistence of the arthritis it causes in humans, no approved vaccines or therapeutic means have been developed for CHIKV infection. Replication of alphaviruses, including CHIKV, is determined not only by their nonstructural proteins but also by a wide range of host factors, which are indispensable components of viral replication complexes (vRCs). Alphavirus nsP3s contain hypervariable domains (HVDs), which encode multiple motifs that drive recruitment of cell- and virus-specific host proteins into vRCs. Our previous data suggested that NAP1 family members are a group of host factors that may interact with CHIKV nsP3 HVD. In this study, we performed a detailed investigation of the NAP1 function in CHIKV replication in vertebrate cells. Our data demonstrate that (i) the NAP1-HVD interactions have strong stimulatory effects on CHIKV replication, (ii) both NAP1L1 and NAP1L4 interact with the CHIKV HVD, (iii) NAP1 family members interact with two motifs, which are located upstream and downstream of the G3BP-binding motifs of CHIKV HVD, (iv) NAP1 proteins interact only with a phosphorylated form of CHIKV HVD, and HVD phosphorylation is mediated by CK2 kinase, and (v) NAP1 and other families of host factors redundantly promote CHIKV replication and their bindings have additive stimulatory effects on viral replication. IMPORTANCE Cellular proteins play critical roles in the assembly of alphavirus replication complexes (vRCs). Their recruitment is determined by the viral nonstructural protein 3 (nsP3). This protein contains a long, disordered hypervariable domain (HVD), which encodes virus-specific combinations of short linear motifs interacting with host factors during vRC assembly. Our study defined the binding mechanism of NAP1 family members to CHIKV HVD and demonstrated a stimulatory effect of this interaction on viral replication. We show that interaction with NAP1L1 is mediated by two HVD motifs and requires phosphorylation of HVD by CK2 kinase. Based on the accumulated data, we present a map of the binding motifs of the critical host factors currently known to interact with CHIKV HVD. It can be used to manipulate cell specificity of viral replication and pathogenesis, and to develop a new generation of vaccine candidates.
Keywords: CK2 kinase; NAP1L1; NAP1L4; alphavirus; chikungunya virus; intrinsically disordered proteins; nsP3; protein phosphorylation; viral pathogenesis; viral replication.
Figures




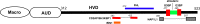
References
Publication types
MeSH terms
Substances
Grants and funding
- R01AI133159/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI118867/AI/NIAID NIH HHS/United States
- R21 AI146969/AI/NIAID NIH HHS/United States
- R01 AI133159/AI/NIAID NIH HHS/United States
- R21AI146969/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

