Antibodies from Rabbits Immunized with HIV-1 Clade B SOSIP Trimers Can Neutralize Multiple Clade B Viruses by Destabilizing the Envelope Glycoprotein
- PMID: 34076487
- PMCID: PMC8354326
- DOI: 10.1128/JVI.00094-21
Antibodies from Rabbits Immunized with HIV-1 Clade B SOSIP Trimers Can Neutralize Multiple Clade B Viruses by Destabilizing the Envelope Glycoprotein
Abstract
The high viral diversity of HIV-1 is a formidable hurdle for the development of an HIV-1 vaccine. Elicitation of broadly neutralizing antibodies (bNAbs) would offer a solution, but so far immunization strategies have failed to efficiently elicit bNAbs. To overcome these obstacles, it is important to understand the immune responses elicited by current HIV-1 envelope glycoprotein (Env) immunogens. To gain more insight, we characterized monoclonal antibodies (MAbs) isolated from rabbits immunized with Env SOSIP trimers based on the clade B isolate AMC008. Four rabbits that were immunized three times with AMC008 trimer developed robust autologous and sporadic low-titer heterologous neutralizing responses. Seventeen AMC008 trimer-reactive MAbs were isolated using antigen-specific single B-cell sorting. Four of these MAbs neutralized the autologous AMC008 virus and several other clade B viruses. When visualized by electron microscopy, the complex of the neutralizing MAbs with the AMC008 trimer showed binding to the gp41 subunit with unusual approach angles, and we observed that their neutralization ability depended on their capacity to induce Env trimer dissociation. Thus, AMC008 SOSIP trimer immunization induced clade B-neutralizing MAbs with unusual approach angles with neutralizing effects that involve trimer destabilization. Optimizing these responses might provide an avenue to the induction of trimer-dissociating bNAbs. IMPORTANCE Roughly 32 million people have died as a consequence of HIV-1 infection since the start of the epidemic, and 1.7 million people still get infected with HIV-1 annually. Therefore, a vaccine to prevent HIV-1 infection is urgently needed. Current HIV-1 immunogens are not able to elicit the broad immune responses needed to provide protection against the large variation of HIV-1 strains circulating globally. A better understanding of the humoral immune responses elicited by immunization with state-of-the-art HIV-1 immunogens should facilitate the design of improved HIV-1 vaccine candidates. We identified antibodies with the ability to neutralize multiple HIV-1 viruses by destabilization of the envelope glycoprotein. Their weak but consistent cross-neutralization ability indicates the potential of this epitope to elicit broad responses. The trimer-destabilizing effect of the neutralizing MAbs, combined with detailed characterization of the neutralization epitope, can be used to shape the next generation of HIV-1 immunogens to elicit improved humoral responses after vaccination.
Keywords: AMC008 SOSIP; HIV-1; approach angle; human immunodeficiency virus; monoclonal antibodies; trimer destabilization; vaccine.
Figures

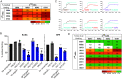


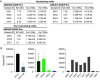

Similar articles
-
Neutralizing Antibody Responses Induced by HIV-1 Envelope Glycoprotein SOSIP Trimers Derived from Elite Neutralizers.J Virol. 2020 Nov 23;94(24):e01214-20. doi: 10.1128/JVI.01214-20. Print 2020 Nov 23. J Virol. 2020. PMID: 32999024 Free PMC article.
-
Antibody responses induced by SHIV infection are more focused than those induced by soluble native HIV-1 envelope trimers in non-human primates.PLoS Pathog. 2021 Aug 25;17(8):e1009736. doi: 10.1371/journal.ppat.1009736. eCollection 2021 Aug. PLoS Pathog. 2021. PMID: 34432859 Free PMC article.
-
HIV-1 Cross-Reactive Primary Virus Neutralizing Antibody Response Elicited by Immunization in Nonhuman Primates.J Virol. 2017 Oct 13;91(21):e00910-17. doi: 10.1128/JVI.00910-17. Print 2017 Nov 1. J Virol. 2017. PMID: 28835491 Free PMC article.
-
Stabilizing HIV-1 envelope glycoprotein trimers to induce neutralizing antibodies.Retrovirology. 2018 Sep 12;15(1):63. doi: 10.1186/s12977-018-0445-y. Retrovirology. 2018. PMID: 30208933 Free PMC article. Review.
-
HIV-1 Envelope Trimer Design and Immunization Strategies To Induce Broadly Neutralizing Antibodies.Trends Immunol. 2016 Mar;37(3):221-232. doi: 10.1016/j.it.2016.01.007. Epub 2016 Feb 9. Trends Immunol. 2016. PMID: 26869204 Free PMC article. Review.
Cited by
-
Fine-mapping the immunodominant antibody epitopes on consensus sequence-based HIV-1 envelope trimer vaccine candidates.NPJ Vaccines. 2022 Nov 25;7(1):152. doi: 10.1038/s41541-022-00576-9. NPJ Vaccines. 2022. PMID: 36433972 Free PMC article.
-
A single residue switch mediates the broad neutralization of Rotaviruses.Nat Commun. 2025 Jan 20;16(1):838. doi: 10.1038/s41467-025-56114-3. Nat Commun. 2025. PMID: 39833145 Free PMC article.
-
Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.PLoS Pathog. 2022 Nov 17;18(11):e1010945. doi: 10.1371/journal.ppat.1010945. eCollection 2022 Nov. PLoS Pathog. 2022. PMID: 36395347 Free PMC article.
-
Lassa virus glycoprotein nanoparticles elicit neutralizing antibody responses and protection.Cell Host Microbe. 2022 Dec 14;30(12):1759-1772.e12. doi: 10.1016/j.chom.2022.10.018. Epub 2022 Nov 17. Cell Host Microbe. 2022. PMID: 36400021 Free PMC article.
-
CoPoP liposomes displaying stabilized clade C HIV-1 Env elicit tier 2 multiclade neutralization in rabbits.Nat Commun. 2024 Apr 11;15(1):3128. doi: 10.1038/s41467-024-47492-1. Nat Commun. 2024. PMID: 38605096 Free PMC article.
References
-
- Dubrovskaya V, Tran K, Ozorowski G, Guenaga J, Wilson R, Bale S, Cottrell CA, Turner HL, Seabright G, O’Dell S, Torres JL, Yang L, Feng Y, Leaman DP, Vázquez Bernat N, Liban T, Louder M, McKee K, Bailer RT, Movsesyan A, Doria-Rose NA, Pancera M, Karlsson Hedestam GB, Zwick MB, Crispin M, Mascola JR, Ward AB, Wyatt RT. 2019. Vaccination with glycan-modified HIV NFL envelope trimer-liposomes elicits broadly neutralizing antibodies to multiple sites of vulnerability. Immunity 51:915–929.e7. 10.1016/j.immuni.2019.10.008. - DOI - PMC - PubMed
-
- Brouwer PJM, Antanasijevic A, Berndsen Z, Yasmeen A, Fiala B, Bijl TPL, Bontjer I, Bale JB, Sheffler W, Allen JD, Schorcht A, Burger JA, Camacho M, Ellis D, Cottrell CA, Behrens AJ, Catalano M, del Moral-Sánchez I, Ketas TJ, LaBranche C, van Gils MJ, Sliepen K, Stewart LJ, Crispin M, Montefiori DC, Baker D, Moore JP, Klasse PJ, Ward AB, King NP, Sanders RW. 2019. Enhancing and shaping the immunogenicity of native-like HIV-1 envelope trimers with a two-component protein nanoparticle. Nat Commun 10:4272. 10.1038/s41467-019-12080-1. - DOI - PMC - PubMed
-
- Rutten L, Lai YT, Blokland S, Truan D, Bisschop IJM, Strokappe NM, Koornneef A, van Manen D, Chuang GY, Farney SK, Schuitemaker H, Kwong PD, Langedijk JPM. 2018. A Universal approach to optimize the folding and stability of prefusion-closed HIV-1 envelope trimers. Cell Rep 23:584–595. 10.1016/j.celrep.2018.03.061. - DOI - PMC - PubMed
-
- Derking R, Ozorowski G, Sliepen K, Yasmeen A, Cupo A, Torres JL, Julien JP, Lee JH, van Montfort T, de Taeye SW, Connors M, Burton DR, Wilson IA, Klasse PJ, Ward AB, Moore JP, Sanders RW. 2015. Comprehensive antigenic map of a cleaved soluble HIV-1 envelope trimer. PLoS Pathog 11:e1004767-22. 10.1371/journal.ppat.1004767. - DOI - PMC - PubMed
-
- Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 76:8875–8889. 10.1128/jvi.76.17.8875-8889.2002. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

