Insights into the Organization of the Poxvirus Multicomponent Entry-Fusion Complex from Proximity Analyses in Living Infected Cells
- PMID: 34076488
- PMCID: PMC8312868
- DOI: 10.1128/JVI.00852-21
Insights into the Organization of the Poxvirus Multicomponent Entry-Fusion Complex from Proximity Analyses in Living Infected Cells
Abstract
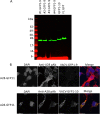
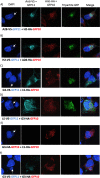
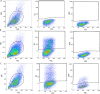
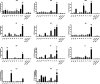
Poxviruses are exceptional in having a complex entry-fusion complex (EFC) that is comprised of 11 conserved proteins embedded in the membrane of mature virions. However, the detailed architecture is unknown and only a few bimolecular protein interactions have been demonstrated by coimmunoprecipitation from detergent-treated lysates and by cross-linking. Here, we adapted the tripartite split green fluorescent protein (GFP) complementation system in order to analyze EFC protein contacts within living cells. This system employs a detector fragment called GFP1-9 comprised of nine GFP β-strands. To achieve fluorescence, two additional 20-amino-acid fragments called GFP10 and GFP11 attached to interacting proteins are needed, providing the basis for identification of the latter. We constructed a novel recombinant vaccinia virus (VACV-GFP1-9) expressing GFP1-9 under a viral early/late promoter and plasmids with VACV late promoters regulating each of the EFC proteins with GFP10 or GFP11 attached to their ectodomains. GFP fluorescence was detected by confocal microscopy at sites of virion assembly in cells infected with VACV-GFP1-9 and cotransfected with plasmids expressing one EFC-GFP10 and one EFC-GFP11 interacting protein. Flow cytometry provided a quantitative way to determine the interaction of each EFC-GFP10 protein with every other EFC-GFP11 protein in the context of a normal infection in which all viral proteins are synthesized and assembled. Previous EFC protein interactions were confirmed, and new ones were discovered and corroborated by additional methods. Most remarkable was the finding that the small, hydrophobic O3 protein interacted with each of the other EFC proteins. IMPORTANCE Poxviruses are enveloped viruses with a DNA-containing core that enters cells following fusion of viral and host membranes. This essential step is a target for vaccines and therapeutics. The entry-fusion complex (EFC) of poxviruses is unusually complex and comprised of 11 conserved viral proteins. Determination of the structure of the EFC is a prerequisite for understanding the fusion mechanism. Here, we used a tripartite split green fluorescent protein assay to determine the proximity of individual EFC proteins in living cells. A network connecting components of the EFC was derived.
Keywords: green fluorescent protein; membrane proteins; multiprotein complex; proximity analysis; vaccinia virus; virus entry.
Figures



Similar articles
-
Mutations Near the N Terminus of Vaccinia Virus G9 Protein Overcome Restrictions on Cell Entry and Syncytium Formation Imposed by the A56/K2 Fusion Regulatory Complex.J Virol. 2020 May 4;94(10):e00077-20. doi: 10.1128/JVI.00077-20. Print 2020 May 4. J Virol. 2020. PMID: 32132239 Free PMC article.
-
Membrane fusion during poxvirus entry.Semin Cell Dev Biol. 2016 Dec;60:89-96. doi: 10.1016/j.semcdb.2016.07.015. Epub 2016 Jul 14. Semin Cell Dev Biol. 2016. PMID: 27423915 Free PMC article. Review.
-
Vaccinia mature virus fusion regulator A26 protein binds to A16 and G9 proteins of the viral entry fusion complex and dissociates from mature virions at low pH.J Virol. 2012 Apr;86(7):3809-18. doi: 10.1128/JVI.06081-11. Epub 2012 Jan 25. J Virol. 2012. PMID: 22278246 Free PMC article.
-
Sequence-divergent chordopoxvirus homologs of the o3 protein maintain functional interactions with components of the vaccinia virus entry-fusion complex.J Virol. 2012 Feb;86(3):1696-705. doi: 10.1128/JVI.06069-11. Epub 2011 Nov 23. J Virol. 2012. PMID: 22114343 Free PMC article.
-
Poxvirus entry and membrane fusion.Virology. 2006 Jan 5;344(1):48-54. doi: 10.1016/j.virol.2005.09.037. Virology. 2006. PMID: 16364735 Review.
Cited by
-
Human Monkeypox: A Comprehensive Overview of Epidemiology, Pathogenesis, Diagnosis, Treatment, and Prevention Strategies.Pathogens. 2023 Jul 18;12(7):947. doi: 10.3390/pathogens12070947. Pathogens. 2023. PMID: 37513794 Free PMC article. Review.
-
The 2.3 Å Structure of A21, a Protein Component of the Conserved Poxvirus Entry-Fusion Complex.J Mol Biol. 2025 Jun 15;437(12):169097. doi: 10.1016/j.jmb.2025.169097. Epub 2025 Mar 19. J Mol Biol. 2025. PMID: 40118206
-
Cross-reactive immune responses to monkeypox virus induced by MVA vaccination in mice.Virol J. 2023 Jun 19;20(1):126. doi: 10.1186/s12985-023-02085-0. Virol J. 2023. PMID: 37337226 Free PMC article.
-
Elucidation of the Cellular Interactome of African Swine Fever Virus Fusion Proteins and Identification of Potential Therapeutic Targets.Viruses. 2023 Apr 29;15(5):1098. doi: 10.3390/v15051098. Viruses. 2023. PMID: 37243184 Free PMC article.
-
Identification of a Potential Entry-Fusion Complex Based on Sequence Homology of African Swine Fever and Vaccinia Virus.Viruses. 2024 Feb 23;16(3):349. doi: 10.3390/v16030349. Viruses. 2024. PMID: 38543715 Free PMC article.
References
-
- Moss B. 2013. Poxviridae, p 2129–2159. In Knipe DM, Howley PM (ed), Fields virology, vol 2. Lippincott/Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

