Genomic analysis of high copy-number sequences for the targeted detection of Listeria species using a flow-through surveillance system
- PMID: 34076739
- PMCID: PMC8289798
- DOI: 10.1007/s00203-021-02388-2
Genomic analysis of high copy-number sequences for the targeted detection of Listeria species using a flow-through surveillance system
Abstract
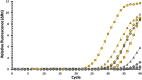
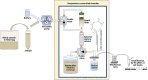
The bacterial foodborne pathogen Listeria monocytogenes has been implicated in fresh produce outbreaks with a significant economic impact. Given that L. monocytogenes is widespread in the environment, food production facilities constantly monitor for the presence of Listeria species. To develop a surveillance platform for food processing facilities, this study conducted a comparative genomic analysis for the identification of conserved high copy sequences in the ribosomal RNA of Listeria species. Simulated folding was performed to assess RNA accessibility in the identified genomic regions targeted for detection, and the developed singleplex assay accurately detected cell amounts lower than 5 cells, while no signals were detected for non-targeted bacteria. The singleplex assay was subsequently tested with a flow-through system, consisting of a DNA aptamer-capture step, followed by sample concentration and mechanical lysis for the detection of Listeria species. Validation experiments indicated the continuous flow-through system accurately detected Listeria species at low cell concentrations.
Keywords: Food safety; Foodborne pathogen; Fresh produce; Genomes; Listeria; RNA.
© 2021. This is a U.S. government work and not under copyright protection in the U.S.; foreign copyright protection may apply.
Conflict of interest statement
Authors BQ, JCY and BGL have no relevant financial or non-financial interests to declare that are relevant to the content of this article. Authors VSDG and DLM are employed by Snap DNA-BioNEMS, Inc. (Mountain View, CA, USA).
Figures




Similar articles
-
Prevalence, identification by a DNA microarray-based assay of human and food isolates Listeria spp. from Tunisia.Pathol Biol (Paris). 2014 Feb;62(1):24-9. doi: 10.1016/j.patbio.2013.10.005. Epub 2014 Jan 23. Pathol Biol (Paris). 2014. PMID: 24461393
-
The VIT technology for rapid detection of Listeria monocytogenes and other Listeria spp.Int J Food Microbiol. 2003 Dec 31;89(2-3):287-90. doi: 10.1016/s0168-1605(03)00307-6. Int J Food Microbiol. 2003. PMID: 14623395
-
Detection of Listeria species and Listeria monocytogenes using polymerase chain reaction.Lett Appl Microbiol. 1990 Sep;11(3):158-62. doi: 10.1111/j.1472-765x.1990.tb00149.x. Lett Appl Microbiol. 1990. PMID: 1367467
-
A new generation of food-borne pathogen detection based on ribosomal RNA.Annu Rev Food Sci Technol. 2013;4:313-25. doi: 10.1146/annurev-food-050412-104448. Annu Rev Food Sci Technol. 2013. PMID: 23464575 Review.
-
A review of the ecology, genomics, and stress response of Listeria innocua and Listeria monocytogenes.Crit Rev Food Sci Nutr. 2012;52(8):712-25. doi: 10.1080/10408398.2010.507909. Crit Rev Food Sci Nutr. 2012. PMID: 22591342 Review.
References
-
- Brouillette R, Aggen D, Borchert B, Buckman K, Domanico M, Fraser-Heaps J, Freier T, Hayman M, Jackson T, Kataoka A, Meyer J, Shoaf E, Stone W. Listeria monocytogenes guidance on environmental monitoring and corrective actions in at-risk foods. Washington: Grocery Manufacturers Association; 2014.
-
- Buchanan RL, Gorris LGM, Hayman MM, Jackson TC, Whiting RC. A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control. 2017;75:1–13. doi: 10.1016/j.foodcont.2016.12.016. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

