Temporal Changes of Virus-Like Particle Abundance and Metagenomic Comparison of Viral Communities in Cropland and Prairie Soils
- PMID: 34077260
- PMCID: PMC8265675
- DOI: 10.1128/mSphere.01160-20
Temporal Changes of Virus-Like Particle Abundance and Metagenomic Comparison of Viral Communities in Cropland and Prairie Soils
Abstract
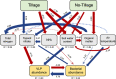
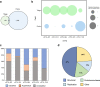
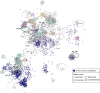
During the last several decades, viruses have been increasingly recognized for their abundance, ubiquity, and important roles in different ecosystems. Despite known contributions to aquatic systems, few studies examine viral abundance and community structure over time in terrestrial ecosystems. The effects of land conversion and land management on soil microbes have been previously investigated, but their effects on virus population are not well studied. This study examined annual dynamics of viral abundance in soils from a native tallgrass prairie and two croplands, conventional till winter wheat and no-till canola, in Oklahoma. Virus-like particle (VLP) abundance varied across sites, and showed clear seasonal shifts. VLP abundance significantly correlated with environmental variables that were generally reflective of land use, including air temperature, soil nitrogen, and plant canopy coverage. Structural equation modeling supported the effects of land use on soil communities by emphasizing interactions between management, environmental factors, and viral and bacterial abundance. Between the viral metagenomes from the prairie and tilled wheat field, 1,231 unique viral operational taxonomic units (vOTUs) were identified, and only five were shared that were rare in the contrasting field. Only 13% of the vOTUs had similarity to previously identified viruses in the RefSeq database, with only 7% having known taxonomic classification. Together, our findings indicated land use and tillage practices influence virus abundance and community structure. Analyses of viromes over time and space are vital to viral ecology in providing insight on viral communities and key information on interactions between viruses, their microbial hosts, and the environment. IMPORTANCE Conversion of land alters the physiochemical and biological environments by not only changing the aboveground community, but also modifying the soil environment for viruses and microbes. Soil microbial communities are critical to nutrient cycling, carbon mineralization, and soil quality; and viruses are known for influencing microbial abundance, community structure, and evolution. Therefore, viruses are considered an important part of soil functions in terrestrial ecosystems. In aquatic environments, virus abundance generally exceeds bacterial counts by an order of magnitude, and they are thought to be one of the greatest genetic reservoirs on the planet. However, data are extremely limited on viruses in soils, and even less is known about their responses to the disturbances associated with land use and management. The study provides important insights into the temporal dynamics of viral abundance and the structure of viral communities in response to the common practice of turning native habitats into arable soils.
Keywords: agriculture; microbial ecology; soil virus; viral ecology; viral metagenomics.
Figures


Similar articles
-
Temporal Dynamics of Bacterial Communities along a Gradient of Disturbance in a U.S. Southern Plains Agroecosystem.mBio. 2022 Jun 28;13(3):e0382921. doi: 10.1128/mbio.03829-21. Epub 2022 Apr 14. mBio. 2022. PMID: 35420482 Free PMC article.
-
Identification of Novel Viruses and Their Microbial Hosts from Soils with Long-Term Nitrogen Fertilization and Cover Cropping Management.mSystems. 2022 Dec 20;7(6):e0057122. doi: 10.1128/msystems.00571-22. Epub 2022 Nov 29. mSystems. 2022. PMID: 36445691 Free PMC article.
-
Soil Viruses Are Underexplored Players in Ecosystem Carbon Processing.mSystems. 2018 Oct 2;3(5):e00076-18. doi: 10.1128/mSystems.00076-18. eCollection 2018 Sep-Oct. mSystems. 2018. PMID: 30320215 Free PMC article.
-
Viruses in Soil Ecosystems: An Unknown Quantity Within an Unexplored Territory.Annu Rev Virol. 2017 Sep 29;4(1):201-219. doi: 10.1146/annurev-virology-101416-041639. Annu Rev Virol. 2017. PMID: 28961409 Review.
-
Counts and sequences, observations that continue to change our understanding of viruses in nature.J Microbiol. 2015 Mar;53(3):181-92. doi: 10.1007/s12275-015-5068-6. Epub 2015 Mar 3. J Microbiol. 2015. PMID: 25732739 Review.
Cited by
-
Viral but not bacterial community successional patterns reflect extreme turnover shortly after rewetting dry soils.Nat Ecol Evol. 2023 Nov;7(11):1809-1822. doi: 10.1038/s41559-023-02207-5. Epub 2023 Sep 28. Nat Ecol Evol. 2023. PMID: 37770548
-
Soil viruses: Understudied agents of soil ecology.Environ Microbiol. 2023 Jan;25(1):143-146. doi: 10.1111/1462-2920.16258. Epub 2022 Nov 5. Environ Microbiol. 2023. PMID: 36271323 Free PMC article. No abstract available.
-
Soil Mycobiome Diversity under Different Tillage Practices in the South of West Siberia.Life (Basel). 2022 Jul 31;12(8):1169. doi: 10.3390/life12081169. Life (Basel). 2022. PMID: 36013348 Free PMC article.
-
Microbial Journey: Mount Everest to Mars.Indian J Microbiol. 2022 Sep;62(3):323-337. doi: 10.1007/s12088-022-01029-6. Epub 2022 Jul 2. Indian J Microbiol. 2022. PMID: 35974919 Free PMC article. Review.
-
Land use conversion increases network complexity and stability of soil microbial communities in a temperate grassland.ISME J. 2023 Dec;17(12):2210-2220. doi: 10.1038/s41396-023-01521-x. Epub 2023 Oct 13. ISME J. 2023. PMID: 37833523 Free PMC article.
References
-
- Liang X, Zhang Y, Wommack KE, Wilhelm SW, DeBruyn JM, Sherfy AC, Zhuang J, Radosevich M. 2020. Lysogenic reproductive strategies of viral communities vary with soil depth and are correlated with bacterial diversity. Soil Biol Biochem 144:107767. doi:10.1016/j.soilbio.2020.107767. - DOI

