The higBA- Type Toxin-Antitoxin System in IncC Plasmids Is a Mobilizable Ciprofloxacin-Inducible System
- PMID: 34077263
- PMCID: PMC8265657
- DOI: 10.1128/mSphere.00424-21
The higBA- Type Toxin-Antitoxin System in IncC Plasmids Is a Mobilizable Ciprofloxacin-Inducible System
Abstract
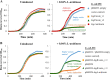
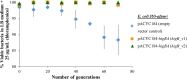
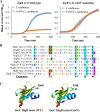
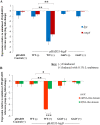
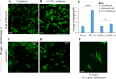
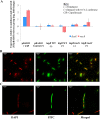
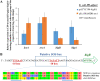
A putative type II toxin-antitoxin (TA) module almost exclusively associated with conjugative IncC plasmids is homologous to the higBA family of TA systems found in chromosomes and plasmids of several species of bacteria. Despite the clinical significance and strong association with high-profile antimicrobial resistance (AMR) genes, the TA system of IncC plasmids remains largely uncharacterized. In this study, we present evidence that IncC plasmids encode a bona fide HigB-like toxin that strongly inhibits bacterial growth and results in cell elongation in Escherichia coli. IncC HigB toxin acts as a ribosome-dependent endoribonuclease that significantly reduces the transcript abundance of a subset of adenine-rich mRNA transcripts. A glycine residue at amino acid position 64 is highly conserved in HigB toxins from different bacterial species, and its replacement with valine (G64V) abolishes the toxicity and the mRNA cleavage activity of the IncC HigB toxin. The IncC plasmid higBA TA system functions as an effective addiction module that maintains plasmid stability in an antibiotic-free environment. This higBA addiction module is the only TA system that we identified in the IncC backbone and appears essential for the stable maintenance of IncC plasmids. We also observed that exposure to subinhibitory concentrations of ciprofloxacin, a DNA-damaging fluoroquinolone antibiotic, results in elevated higBA expression, which raises interesting questions about its regulatory mechanisms. A better understanding of this higBA-type TA module potentially allows for its subversion as part of an AMR eradication strategy. IMPORTANCE Toxin-antitoxin (TA) systems play vital roles in maintaining plasmids in bacteria. Plasmids with incompatibility group C are large plasmids that disseminate via conjugation and carry high-profile antibiotic resistance genes. We present experimental evidence that IncC plasmids carry a TA system that functions as an effective addiction module and maintains plasmid stability in an antibiotic-free environment. The toxin of IncC plasmids acts as an endoribonuclease that targets a subset of mRNA transcripts. Overexpressing the IncC toxin gene strongly inhibits bacterial growth and results in cell elongation in Escherichia coli hosts. We also identify a conserved amino acid residue in the toxin protein that is essential for its toxicity and show that the expression of this TA system is activated by a DNA-damaging antibiotic, ciprofloxacin. This mobile TA system may contribute to managing bacterial stress associated with DNA-damaging antibiotics.
Keywords: Enterobacteriaceae; antibiotic resistance; plasmids; toxin-antitoxin systems.
Figures

Similar articles
-
A noncanonical intrinsic terminator in the HicAB toxin-antitoxin operon promotes the transmission of conjugative antibiotic resistance plasmids.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf125. doi: 10.1093/nar/gkaf125. Nucleic Acids Res. 2025. PMID: 40036506 Free PMC article.
-
Shared mechanisms of enhanced plasmid maintenance and antibiotic tolerance mediated by the VapBC toxin:antitoxin system.mBio. 2025 Feb 5;16(2):e0261624. doi: 10.1128/mbio.02616-24. Epub 2024 Dec 20. mBio. 2025. PMID: 39704502 Free PMC article.
-
2.09 Å Resolution structure of E. coli HigBA toxin-antitoxin complex reveals an ordered DNA-binding domain and intrinsic dynamics in antitoxin.Biochem J. 2020 Oct 30;477(20):4001-4019. doi: 10.1042/BCJ20200363. Biochem J. 2020. PMID: 33000860
-
[Functions of bacterial Toxin-Antitoxin systems].Sheng Wu Gong Cheng Xue Bao. 2018 Aug 25;34(8):1270-1278. doi: 10.13345/j.cjb.170527. Sheng Wu Gong Cheng Xue Bao. 2018. PMID: 30152212 Review. Chinese.
-
Structural Variations and Rearrangements in Bacterial Type II Toxin-Antitoxin Systems.Subcell Biochem. 2024;104:245-267. doi: 10.1007/978-3-031-58843-3_11. Subcell Biochem. 2024. PMID: 38963490 Review.
Cited by
-
Biological Functions of Type II Toxin-Antitoxin Systems in Bacteria.Microorganisms. 2021 Jun 11;9(6):1276. doi: 10.3390/microorganisms9061276. Microorganisms. 2021. PMID: 34208120 Free PMC article. Review.
-
Novel insights into genetic characteristics of blaGES-encoding plasmids from hospital sewage.Front Microbiol. 2023 Aug 17;14:1209195. doi: 10.3389/fmicb.2023.1209195. eCollection 2023. Front Microbiol. 2023. PMID: 37664110 Free PMC article.
-
A Novel Plasmid Entry Exclusion System in pKPC_UVA01, a Promiscuous Conjugative Plasmid Carrying the blaKPC Carbapenemase Gene.Antimicrob Agents Chemother. 2022 Mar 15;66(3):e0232221. doi: 10.1128/aac.02322-21. Epub 2022 Jan 10. Antimicrob Agents Chemother. 2022. PMID: 35007138 Free PMC article.
-
Epidemiological and biological characteristics of IncR plasmids as multihost antibiotic resistance carriers.Front Microbiol. 2025 Jul 17;16:1622352. doi: 10.3389/fmicb.2025.1622352. eCollection 2025. Front Microbiol. 2025. PMID: 40746312 Free PMC article. Review.
-
Two birds with one stone: SGI1 can stabilize itself and expel the IncC helper by hijacking the plasmid parABS system.Nucleic Acids Res. 2024 Mar 21;52(5):2498-2518. doi: 10.1093/nar/gkae050. Nucleic Acids Res. 2024. PMID: 38300764 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

