The nonreceptor tyrosine kinase SRMS inhibits autophagy and promotes tumor growth by phosphorylating the scaffolding protein FKBP51
- PMID: 34077419
- PMCID: PMC8202955
- DOI: 10.1371/journal.pbio.3001281
The nonreceptor tyrosine kinase SRMS inhibits autophagy and promotes tumor growth by phosphorylating the scaffolding protein FKBP51
Abstract
Nutrient-responsive protein kinases control the balance between anabolic growth and catabolic processes such as autophagy. Aberrant regulation of these kinases is a major cause of human disease. We report here that the vertebrate nonreceptor tyrosine kinase Src-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites (SRMS) inhibits autophagy and promotes growth in a nutrient-responsive manner. Under nutrient-replete conditions, SRMS phosphorylates the PHLPP scaffold FK506-binding protein 51 (FKBP51), disrupts the FKBP51-PHLPP complex, and promotes FKBP51 degradation through the ubiquitin-proteasome pathway. This prevents PHLPP-mediated dephosphorylation of AKT, causing sustained AKT activation that promotes growth and inhibits autophagy. SRMS is amplified and overexpressed in human cancers where it drives unrestrained AKT signaling in a kinase-dependent manner. SRMS kinase inhibition activates autophagy, inhibits cancer growth, and can be accomplished using the FDA-approved tyrosine kinase inhibitor ibrutinib. This illuminates SRMS as a targetable vulnerability in human cancers and as a new target for pharmacological induction of autophagy in vertebrates.
Conflict of interest statement
I have read he journal’s policy and the authors of this manuscript have the following competing interests: J.M.P., S.W.Y., and M.B.P. are employees and shareholders of Amgen. Author Sergei von Hoyningen-Huene was unavailable to confirm their authorship contributions. On their behalf, the corresponding author has reported their contributions to the best of their knowledge.
Figures
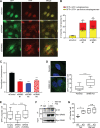
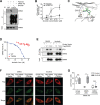
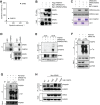

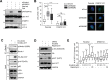
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

