Neoplastic and immune single-cell transcriptomics define subgroup-specific intra-tumoral heterogeneity of childhood medulloblastoma
- PMID: 34077540
- PMCID: PMC8804892
- DOI: 10.1093/neuonc/noab135
Neoplastic and immune single-cell transcriptomics define subgroup-specific intra-tumoral heterogeneity of childhood medulloblastoma
Abstract
Background: Medulloblastoma (MB) is a heterogeneous disease in which neoplastic cells and associated immune cells contribute to disease progression. We aimed to determine the influence of neoplastic and immune cell diversity on MB biology in patient samples and animal models.
Methods: To better characterize cellular heterogeneity in MB we used single-cell RNA sequencing, immunohistochemistry, and deconvolution of transcriptomic data to profile neoplastic and immune populations in patient samples and animal models across childhood MB subgroups.
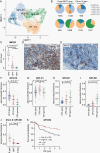
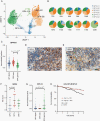
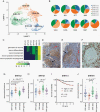
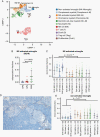
Results: Neoplastic cells cluster primarily according to individual sample of origin which is influenced by chromosomal copy number variance. Harmony alignment reveals novel MB subgroup/subtype-associated subpopulations that recapitulate neurodevelopmental processes, including photoreceptor and glutamatergic neuron-like cells in molecular subgroups GP3 and GP4, and a specific nodule-associated neuronally differentiated subpopulation in the sonic hedgehog subgroup. We definitively chart the spectrum of MB immune cell infiltrates, which include subpopulations that recapitulate developmentally related neuron-pruning and antigen-presenting myeloid cells. MB cellular diversity matching human samples is mirrored in subgroup-specific mouse models of MB.
Conclusions: These findings provide a clearer understanding of the diverse neoplastic and immune cell subpopulations that constitute the MB microenvironment.
Keywords: immune; medulloblastoma; neoplastic; scRNA-seq; subpopulation.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

Comment in
-
Faraway, so close: Intratumoral heterogeneity of medulloblastoma subgroups.Neuro Oncol. 2022 Feb 1;24(2):287-288. doi: 10.1093/neuonc/noab281. Neuro Oncol. 2022. PMID: 34865140 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

