An acquired acyltransferase promotes Klebsiella pneumoniae ST258 respiratory infection
- PMID: 34077733
- PMCID: PMC8283688
- DOI: 10.1016/j.celrep.2021.109196
An acquired acyltransferase promotes Klebsiella pneumoniae ST258 respiratory infection
Abstract
Klebsiella pneumoniae ST258 is a human pathogen associated with poor outcomes worldwide. We identify a member of the acyltransferase superfamily 3 (atf3), enriched within the ST258 clade, that provides a major competitive advantage for the proliferation of these organisms in vivo. Comparison of a wild-type ST258 strain (KP35) and a Δatf3 isogenic mutant generated by CRISPR-Cas9 targeting reveals greater NADH:ubiquinone oxidoreductase transcription and ATP generation, fueled by increased glycolysis. The acquisition of atf3 induces changes in the bacterial acetylome, promoting lysine acetylation of multiple proteins involved in central metabolism, specifically Zwf (glucose-6 phosphate dehydrogenase). The atf3-mediated metabolic boost leads to greater consumption of glucose in the host airway and increased bacterial burden in the lung, independent of cytokine levels and immune cell recruitment. Acquisition of this acyltransferase enhances fitness of a K. pneumoniae ST258 isolate and may contribute to the success of this clonal complex as a healthcare-associated pathogen.
Keywords: Klebsiella pneumoniae; acetylation; bacterial pneumonia; glycolysis; metabolism; sequence type 258.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



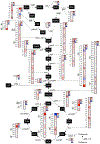


References
-
- Andersson DI, and Hughes D (2010). Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol 8, 260–271. - PubMed
-
- Blin C, Passet V, Touchon M, Rocha EPC, and Brisse S (2017). Metabolic diversity of the emerging pathogenic lineages of Klebsiella pneumoniae. Environ. Microbiol 19, 1881–1898. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

