The reduced genome of a heritable symbiont from an ectoparasitic feather feeding louse
- PMID: 34078265
- PMCID: PMC8173840
- DOI: 10.1186/s12862-021-01840-7
The reduced genome of a heritable symbiont from an ectoparasitic feather feeding louse
Abstract
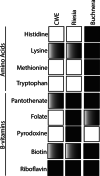
Background: Feather feeding lice are abundant and diverse ectoparasites that complete their entire life cycle on an avian host. The principal or sole source of nutrition for these lice is feathers. Feathers appear to lack four amino acids that the lice would require to complete development and reproduce. Several insect groups have acquired heritable and intracellular bacteria that can synthesize metabolites absent in an insect's diet, allowing insects to feed exclusively on nutrient-poor resources. Multiple species of feather feeding lice have been shown to harbor heritable and intracellular bacteria. We expected that these bacteria augment the louse's diet with amino acids and facilitated the evolution of these diverse and specialized parasites. Heritable symbionts of insects often have small genomes that contain a minimal set of genes needed to maintain essential cell functions and synthesize metabolites absent in the host insect's diet. Therefore, we expected the genome of a bacterial endosymbiont in feather lice would be small, but encode pathways for biosynthesis of amino acids.
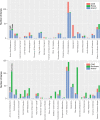
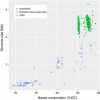
Results: We sequenced the genome of a bacterial symbiont from a feather feeding louse (Columbicola wolffhuegeli) that parasitizes the Pied Imperial Pigeon (Ducula bicolor) and used its genome to predict metabolism of amino acids based on the presence or absence of genes. We found that this bacterial symbiont has a small genome, similar to the genomes of heritable symbionts described in other insect groups. However, we failed to identify many of the genes that we expected would support metabolism of amino acids in the symbiont genome. We also evaluated other gene pathways and features of the highly reduced genome of this symbiotic bacterium.
Conclusions: Based on the data collected in this study, it does not appear that this bacterial symbiont can synthesize amino acids needed to complement the diet of a feather feeding louse. Our results raise additional questions about the biology of feather chewing lice and the roles of symbiotic bacteria in evolution of diverse avian parasites.
Keywords: Endosymbiont; Genome reduction; Keratin; Metabolic complementation; Phthiraptera.
Conflict of interest statement
The authors have not competing interests to declare.
Figures

Similar articles
-
An obligate symbiont of Haematomyzus elephantis with a strongly reduced genome resembles symbiotic bacteria in sucking lice.Appl Environ Microbiol. 2025 Jun 18;91(6):e0022025. doi: 10.1128/aem.00220-25. Epub 2025 May 14. Appl Environ Microbiol. 2025. PMID: 40366182 Free PMC article.
-
Two Bacterial Genera, Sodalis and Rickettsia, Associated with the Seal Louse Proechinophthirus fluctus (Phthiraptera: Anoplura).Appl Environ Microbiol. 2016 May 16;82(11):3185-97. doi: 10.1128/AEM.00282-16. Print 2016 Jun 1. Appl Environ Microbiol. 2016. PMID: 26994086 Free PMC article.
-
Primates, Lice and Bacteria: Speciation and Genome Evolution in the Symbionts of Hominid Lice.Mol Biol Evol. 2017 Jul 1;34(7):1743-1757. doi: 10.1093/molbev/msx117. Mol Biol Evol. 2017. PMID: 28419279 Free PMC article.
-
Signatures of host/symbiont genome coevolution in insect nutritional endosymbioses.Proc Natl Acad Sci U S A. 2015 Aug 18;112(33):10255-61. doi: 10.1073/pnas.1423305112. Epub 2015 May 26. Proc Natl Acad Sci U S A. 2015. PMID: 26039986 Free PMC article. Review.
-
The evolutionary development of plant-feeding insects and their nutritional endosymbionts.Insect Sci. 2017 Dec;24(6):910-928. doi: 10.1111/1744-7917.12463. Epub 2017 May 30. Insect Sci. 2017. PMID: 28371395 Review.
Cited by
-
Stochasticity, determinism, and contingency shape genome evolution of endosymbiotic bacteria.Nat Commun. 2024 May 29;15(1):4571. doi: 10.1038/s41467-024-48784-2. Nat Commun. 2024. PMID: 38811551 Free PMC article.
-
Phylogenetic relationship between the endosymbiont "Candidatus Riesia pediculicola" and its human louse host.Parasit Vectors. 2022 Mar 5;15(1):73. doi: 10.1186/s13071-022-05203-z. Parasit Vectors. 2022. PMID: 35248159 Free PMC article.
-
An obligate symbiont of Haematomyzus elephantis with a strongly reduced genome resembles symbiotic bacteria in sucking lice.Appl Environ Microbiol. 2025 Jun 18;91(6):e0022025. doi: 10.1128/aem.00220-25. Epub 2025 May 14. Appl Environ Microbiol. 2025. PMID: 40366182 Free PMC article.
-
Supergroup F Wolbachia with extremely reduced genome: transition to obligate insect symbionts.Microbiome. 2023 Feb 7;11(1):22. doi: 10.1186/s40168-023-01462-9. Microbiome. 2023. PMID: 36750860 Free PMC article.
-
Genomic Approaches to Uncovering the Coevolutionary History of Parasitic Lice.Life (Basel). 2022 Sep 16;12(9):1442. doi: 10.3390/life12091442. Life (Basel). 2022. PMID: 36143478 Free PMC article. Review.
References
-
- Davis GR. Essential dietary amino acids for growth of larvae of the yellow mealworm Tenebrio molitor L. J Nutr. 1975;1051071:1075. - PubMed
Publication types
MeSH terms
Grants and funding
- DEB-1239788/Virginia Commonwealth University Life Sciences and National Science Foundation awards
- DEB-1342604/Virginia Commonwealth University Life Sciences and National Science Foundation awards
- DEB-1855812/Virginia Commonwealth University Life Sciences and National Science Foundation awards
- DEB-1926919/Virginia Commonwealth University Life Sciences and National Science Foundation awards
LinkOut - more resources
Full Text Sources
