Specific N-cadherin-dependent pathways drive human breast cancer dormancy in bone marrow
- PMID: 34078741
- PMCID: PMC8200294
- DOI: 10.26508/lsa.202000969
Specific N-cadherin-dependent pathways drive human breast cancer dormancy in bone marrow
Abstract
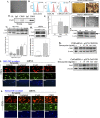
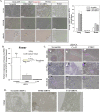
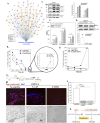
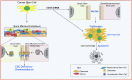
The challenge for treating breast cancer (BC) is partly due to long-term dormancy driven by cancer stem cells (CSCs) capable of evading immune response and resist chemotherapy. BC cells show preference for the BM, resulting in poor prognosis. CSCs use connexin 43 (Cx43) to form gap junctional intercellular communication with BM niche cells, fibroblasts, and mesenchymal stem cells (MSCs). However, Cx43 is an unlikely target to reverse BC dormancy because of its role as a hematopoietic regulator. We found N-cadherin (CDH2) and its associated pathways as potential drug targets. CDH2, highly expressed in CSCs, interacts intracellularly with Cx43, colocalizes with Cx43 in BC cells within BM biopsies of patients, and is required for Cx43-mediated gap junctional intercellular communication with BM niche cells. Notably, CDH2 and anti-apoptotic pathways maintained BC dormancy. We thereby propose these pathways as potential pharmacological targets to prevent dormancy and chemosensitize resistant CSCs.
© 2021 Sinha et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures









References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
