Epigenetic reprogramming promotes the antiviral action of IFNα in HBV-infected cells
- PMID: 34078875
- PMCID: PMC8170866
- DOI: 10.1038/s41420-021-00515-y
Epigenetic reprogramming promotes the antiviral action of IFNα in HBV-infected cells
Abstract
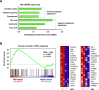
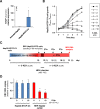
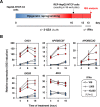
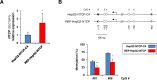
Chronic hepatitis B virus (HBV) infections remain a health burden affecting ~250 million people worldwide. Thus far, available interferon-alpha (IFNα)-based therapies have shown unsatisfactory cure rates, and alternative therapeutic molecules are still required. However, their development has been hampered because accessible cell models supporting relevant HBV replication and appropriate antiviral activity are lacking. Strategies that reverse epigenetic alterations offer a unique opportunity for cell reprogramming, which is valuable for restoring altered cellular functions in human cell lines. This work aimed to investigate the feasibility of converting HepG2 cells that stably overexpress the HBV entry receptor (sodium/taurocholate cotransporting polypeptide, NTCP) toward IFNα-responsive cells using epigenetic reprogramming. Herein, we showed that an epigenetic regimen with non-cytotoxic doses of the demethylating compound 5-azacytidine restored the anti-HBV action of IFNα in epigenetically reprogrammed HepG2-NTCP-C4 cells, named REP-HepG2-NTCP cells. Thus, a significant inhibition in HBV DNA levels was measured in REP-HepG2-NTCP cells after IFNα treatment. This inhibitory effect was associated with the enhancement of IFNα-mediated induction of critical interferon-stimulated genes (ISGs), which was limited in non-reprogrammed cells. In particular, our data indicated that re-expression of 2'-5'-oligoadenylate synthetase 1 (OAS1) and interferon regulatory factor 9 (IRF9) was the result of an epigenetically driven unmasking of these genes in reprogrammed cells. At last, we evaluated the therapeutic potential of the IFN analog CDM-3008 in REP-HepG2-NTCP cells and demonstrated the efficiency of this chemical compound in triggering ISG induction and HBV inhibition. In summary, this study shows that epigenetic reprogramming promotes the IFNα response in HBV-infected cells and is potentially attractive for cell-based experimental screening of IFN-like compounds.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
An interferon-like small chemical compound CDM-3008 suppresses hepatitis B virus through induction of interferon-stimulated genes.PLoS One. 2019 Jun 12;14(6):e0216139. doi: 10.1371/journal.pone.0216139. eCollection 2019. PLoS One. 2019. PMID: 31188831 Free PMC article.
-
Unusual Features of Sodium Taurocholate Cotransporting Polypeptide as a Hepatitis B Virus Receptor.J Virol. 2016 Aug 26;90(18):8302-13. doi: 10.1128/JVI.01153-16. Print 2016 Sep 15. J Virol. 2016. PMID: 27384660 Free PMC article.
-
A new high-content screening assay of the entire hepatitis B virus life cycle identifies novel antivirals.JHEP Rep. 2021 Apr 30;3(4):100296. doi: 10.1016/j.jhepr.2021.100296. eCollection 2021 Aug. JHEP Rep. 2021. PMID: 34222850 Free PMC article.
-
Hepatitis delta virus: insights into a peculiar pathogen and novel treatment options.Nat Rev Gastroenterol Hepatol. 2016 Oct;13(10):580-9. doi: 10.1038/nrgastro.2016.126. Epub 2016 Aug 18. Nat Rev Gastroenterol Hepatol. 2016. PMID: 27534692 Review.
-
HBV Genome and Life Cycle.Adv Exp Med Biol. 2020;1179:17-37. doi: 10.1007/978-981-13-9151-4_2. Adv Exp Med Biol. 2020. PMID: 31741332 Review.
Cited by
-
miRNA-29b-1-5p mediates an antiviral activity by targeting the HBV entry receptor in human hepatocytes.Sci Rep. 2025 Jul 3;15(1):23725. doi: 10.1038/s41598-025-09655-y. Sci Rep. 2025. PMID: 40610739 Free PMC article.
-
Role of Virus-Induced Host Cell Epigenetic Changes in Cancer.Int J Mol Sci. 2021 Aug 3;22(15):8346. doi: 10.3390/ijms22158346. Int J Mol Sci. 2021. PMID: 34361112 Free PMC article. Review.
-
Harnessing Epigenetics: Innovative Approaches in Diagnosing and Combating Viral Acute Respiratory Infections.Pathogens. 2025 Feb 1;14(2):129. doi: 10.3390/pathogens14020129. Pathogens. 2025. PMID: 40005506 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

