Confirming putative variants at ≤ 5% allele frequency using allele enrichment and Sanger sequencing
- PMID: 34079006
- PMCID: PMC8172533
- DOI: 10.1038/s41598-021-91142-1
Confirming putative variants at ≤ 5% allele frequency using allele enrichment and Sanger sequencing
Abstract
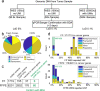
Whole exome sequencing (WES) is used to identify mutations in a patient's tumor DNA that are predictive of tumor behavior, including the likelihood of response or resistance to cancer therapy. WES has a mutation limit of detection (LoD) at variant allele frequencies (VAF) of 5%. Putative mutations called at ≤ 5% VAF are frequently due to sequencing errors, therefore reporting these subclonal mutations incurs risk of significant false positives. Here we performed ~ 1000 × WES on fresh-frozen and formalin-fixed paraffin-embedded (FFPE) tissue biopsy samples from a non-small cell lung cancer patient, and identified 226 putative mutations at between 0.5 and 5% VAF. Each variant was then tested using NuProbe NGSure, to confirm the original WES calls. NGSure utilizes Blocker Displacement Amplification to first enrich the allelic fraction of the mutation and then uses Sanger sequencing to determine mutation identity. Results showed that 52% of the 226 (117) putative variants were disconfirmed, among which 2% (5) putative variants were found to be misidentified in WES. In the 66 cancer-related variants, the disconfirmed rate was 82% (54/66). This data demonstrates Blocker Displacement Amplification allelic enrichment coupled with Sanger sequencing can be used to confirm putative mutations ≤ 5% VAF. By implementing this method, next-generation sequencing can reliably report low-level variants at a high sensitivity, without the cost of high sequencing depth.
Conflict of interest statement
S.X.C. and L.Y.C. consult for NuProbe USA. D.Y.Z. declares a competing interest in the form of consulting for and equity ownership in NuProbe USA, Torus Biosystems, and Pana Bio. All other authors declare no conflict of interest.
Figures



References
-
- Lier, A. et al. Validating comprehensive next-generation sequencing results for precision oncology: The NCT/DKTK molecularly aided stratification for tumor eradication research experience. JCO Precis. Oncol., 1–13 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

