NOTUM from Apc-mutant cells biases clonal competition to initiate cancer
- PMID: 34079124
- PMCID: PMC7615049
- DOI: 10.1038/s41586-021-03525-z
NOTUM from Apc-mutant cells biases clonal competition to initiate cancer
Abstract
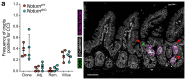
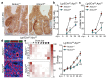
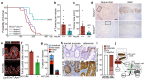
The tumour suppressor APC is the most commonly mutated gene in colorectal cancer. Loss of Apc in intestinal stem cells drives the formation of adenomas in mice via increased WNT signalling1, but reduced secretion of WNT ligands increases the ability of Apc-mutant intestinal stem cells to colonize a crypt (known as fixation)2. Here we investigated how Apc-mutant cells gain a clonal advantage over wild-type counterparts to achieve fixation. We found that Apc-mutant cells are enriched for transcripts that encode several secreted WNT antagonists, with Notum being the most highly expressed. Conditioned medium from Apc-mutant cells suppressed the growth of wild-type organoids in a NOTUM-dependent manner. Furthermore, NOTUM-secreting Apc-mutant clones actively inhibited the proliferation of surrounding wild-type crypt cells and drove their differentiation, thereby outcompeting crypt cells from the niche. Genetic or pharmacological inhibition of NOTUM abrogated the ability of Apc-mutant cells to expand and form intestinal adenomas. We identify NOTUM as a key mediator during the early stages of mutation fixation that can be targeted to restore wild-type cell competitiveness and provide preventative strategies for people at a high risk of developing colorectal cancer.
Conflict of interest statement
The authors have no competing interests to declare.
Figures









Comment in
-
Cancer stem cells in the gut have a bad influence on neighbouring cells.Nature. 2021 Jun;594(7863):340-341. doi: 10.1038/d41586-021-01379-z. Nature. 2021. PMID: 34079113 No abstract available.
-
Holding back the competition.Nat Rev Cancer. 2021 Aug;21(8):480. doi: 10.1038/s41568-021-00384-8. Nat Rev Cancer. 2021. PMID: 34172964 No abstract available.
-
Race to the bottom: Darwinian competition in early intestinal tumorigenesis.Cell Stem Cell. 2021 Aug 5;28(8):1340-1342. doi: 10.1016/j.stem.2021.07.008. Cell Stem Cell. 2021. PMID: 34358438
-
Outcompeting neighbors for intestinal cancer initiation.Nat Cancer. 2021 Dec;2(12):1292. doi: 10.1038/s43018-021-00314-5. Nat Cancer. 2021. PMID: 35121927 No abstract available.
References
-
- Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

