Tracing oncogene-driven remodelling of the intestinal stem cell niche
- PMID: 34079126
- PMCID: PMC7614896
- DOI: 10.1038/s41586-021-03605-0
Tracing oncogene-driven remodelling of the intestinal stem cell niche
Abstract
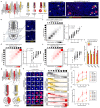
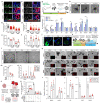
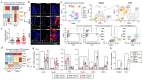
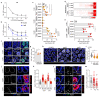
Interactions between tumour cells and the surrounding microenvironment contribute to tumour progression, metastasis and recurrence1-3. Although mosaic analyses in Drosophila have advanced our understanding of such interactions4,5, it has been difficult to engineer parallel approaches in vertebrates. Here we present an oncogene-associated, multicolour reporter mouse model-the Red2Onco system-that allows differential tracing of mutant and wild-type cells in the same tissue. By applying this system to the small intestine, we show that oncogene-expressing mutant crypts alter the cellular organization of neighbouring wild-type crypts, thereby driving accelerated clonal drift. Crypts that express oncogenic KRAS or PI3K secrete BMP ligands that suppress local stem cell activity, while changes in PDGFRloCD81+ stromal cells induced by crypts with oncogenic PI3K alter the WNT signalling environment. Together, these results show how oncogene-driven paracrine remodelling creates a niche environment that is detrimental to the maintenance of wild-type tissue, promoting field transformation dominated by oncogenic clones.
Conflict of interest statement
The authors declare no competing interests.
Figures











Comment in
-
Cancer stem cells in the gut have a bad influence on neighbouring cells.Nature. 2021 Jun;594(7863):340-341. doi: 10.1038/d41586-021-01379-z. Nature. 2021. PMID: 34079113 No abstract available.
-
Holding back the competition.Nat Rev Cancer. 2021 Aug;21(8):480. doi: 10.1038/s41568-021-00384-8. Nat Rev Cancer. 2021. PMID: 34172964 No abstract available.
-
Race to the bottom: Darwinian competition in early intestinal tumorigenesis.Cell Stem Cell. 2021 Aug 5;28(8):1340-1342. doi: 10.1016/j.stem.2021.07.008. Cell Stem Cell. 2021. PMID: 34358438
-
Outcompeting neighbors for intestinal cancer initiation.Nat Cancer. 2021 Dec;2(12):1292. doi: 10.1038/s43018-021-00314-5. Nat Cancer. 2021. PMID: 35121927 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

