Cell Type-Specific Differentiation Between Indica and Japonica Rice Root Tip Responses to Different Environments Based on Single-Cell RNA Sequencing
- PMID: 34079581
- PMCID: PMC8166412
- DOI: 10.3389/fgene.2021.659500
Cell Type-Specific Differentiation Between Indica and Japonica Rice Root Tip Responses to Different Environments Based on Single-Cell RNA Sequencing
Abstract
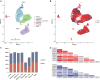
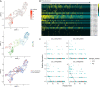
Background: As Oryza sativa ssp. indica and Oryza sativa ssp. japonica are the two major subspecies of Asian cultivated rice, the adaptative evolution of these varieties in divergent environments is an important topic in both theoretical and practical studies. However, the cell type-specific differentiation between indica and japonica rice varieties in response to divergent habitat environments, which facilitates an understanding of the genetic basis underlying differentiation and environmental adaptation between rice subspecies at the cellular level, is little known. Methods: We analyzed a published single-cell RNA sequencing dataset to explore the differentially expressed genes between indica and japonica rice varieties in each cell type. To estimate the relationship between cell type-specific differentiation and environmental adaptation, we focused on genes in the WRKY, NAC, and BZIP transcription factor families, which are closely related to abiotic stress responses. In addition, we integrated five bulk RNA sequencing datasets obtained under conditions of abiotic stress, including cold, drought and salinity, in this study. Furthermore, we analyzed quiescent center cells in rice root tips based on orthologous markers in Arabidopsis. Results: We found differentially expressed genes between indica and japonica rice varieties with cell type-specific patterns, which were enriched in the pathways related to root development and stress reposes. Some of these genes were members of the WRKY, NAC, and BZIP transcription factor families and were differentially expressed under cold, drought or salinity stress. In addition, LOC_Os01g16810, LOC_Os01g18670, LOC_Os04g52960, and LOC_Os08g09350 may be potential markers of quiescent center cells in rice root tips. Conclusion: These results identified cell type-specific differentially expressed genes between indica-japonica rice varieties that were related to various environmental stresses and provided putative markers of quiescent center cells. This study provides new clues for understanding the development and physiology of plants during the process of adaptative divergence, in addition to identifying potential target genes for the improvement of stress tolerance in rice breeding applications.
Keywords: Oryza sativa; adaptative evolution; indica-japonica differentiation; quiescent center cells; single-cell RNA sequencing; transcription factor family.
Copyright © 2021 Wang, Cheng, Fan, Zhang, Zhang and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Microsatellite diversity within Oryza sativa with emphasis on indica-japonica divergence.Genet Res. 2005 Feb;85(1):1-14. doi: 10.1017/s0016672304007293. Genet Res. 2005. PMID: 16089032
-
Differential alternative polyadenylation contributes to the developmental divergence between two rice subspecies, japonica and indica.Plant J. 2019 Apr;98(2):260-276. doi: 10.1111/tpj.14209. Epub 2019 Feb 13. Plant J. 2019. PMID: 30570805
-
Genetic dissection of eating and cooking qualities in different subpopulations of cultivated rice (Oryza sativa L.) through association mapping.BMC Genet. 2020 Oct 14;21(1):119. doi: 10.1186/s12863-020-00922-7. BMC Genet. 2020. PMID: 33054745 Free PMC article.
-
Advances on the Study of Diurnal Flower-Opening Times of Rice.Int J Mol Sci. 2023 Jun 26;24(13):10654. doi: 10.3390/ijms241310654. Int J Mol Sci. 2023. PMID: 37445832 Free PMC article. Review.
-
Introgression among subgroups is an important driving force for genetic improvement and evolution of the Asian cultivated rice Oryza sativa L.Front Plant Sci. 2025 Feb 20;16:1535880. doi: 10.3389/fpls.2025.1535880. eCollection 2025. Front Plant Sci. 2025. PMID: 40051880 Free PMC article. Review.
Cited by
-
A single-cell and spatial wheat root atlas with cross-species annotations delineates conserved tissue-specific marker genes and regulators.Cell Rep. 2025 Feb 25;44(2):115240. doi: 10.1016/j.celrep.2025.115240. Epub 2025 Feb 1. Cell Rep. 2025. PMID: 39893633 Free PMC article.
-
Accelerating crop improvement via integration of transcriptome-based network biology and genome editing.Planta. 2025 Mar 17;261(4):92. doi: 10.1007/s00425-025-04666-5. Planta. 2025. PMID: 40095140 Review.
-
Single-cell transcriptome sequencing reveals the sphingolipid metabolism pathway plays an important role in leaf senescence in tobacco.BMC Plant Biol. 2025 Aug 6;25(1):1026. doi: 10.1186/s12870-025-07051-2. BMC Plant Biol. 2025. PMID: 40764913 Free PMC article.
-
Opportunities and challenges in the application of single-cell transcriptomics in plant tissue research.Physiol Mol Biol Plants. 2025 Feb;31(2):199-209. doi: 10.1007/s12298-025-01558-6. Epub 2025 Feb 22. Physiol Mol Biol Plants. 2025. PMID: 40070535 Review.
-
Surviving a Double-Edged Sword: Response of Horticultural Crops to Multiple Abiotic Stressors.Int J Mol Sci. 2024 May 10;25(10):5199. doi: 10.3390/ijms25105199. Int J Mol Sci. 2024. PMID: 38791235 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials

