The pleiotropic roles of circular and long noncoding RNAs in cutaneous melanoma
- PMID: 34080276
- PMCID: PMC8807361
- DOI: 10.1002/1878-0261.13034
The pleiotropic roles of circular and long noncoding RNAs in cutaneous melanoma
Abstract
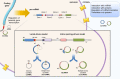
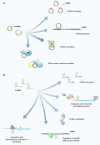
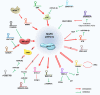
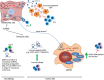
Cutaneous melanoma (CM) is a very aggressive disease, often characterized by unresponsiveness to conventional therapies and high mortality rates worldwide. The identification of the activating BRAFV600 mutations in approximately 50% of CM patients has recently fueled the development of novel small-molecule inhibitors that specifically target BRAFV600 -mutant CM. In addition, a major progress in CM treatment has been made by monoclonal antibodies that regulate the immune checkpoint inhibitors. However, although target-based therapies and immunotherapeutic strategies have yielded promising results, CM treatment remains a major challenge. In the last decade, accumulating evidence points to the aberrant expression of different types of noncoding RNAs (ncRNAs) in CM. While studies on microRNAs have grown exponentially leading to significant insights on CM biology, the role of circular RNAs (circRNAs) and long noncoding RNAs (lncRNAs) in this tumor is less understood, and much remains to be discovered. Here, we summarize and critically review the available evidence on the molecular functions of circRNAs and lncRNAs in BRAFV600 -mutant CM and CM immunogenicity, providing recent updates on their functional role in targeted therapy and immunotherapy resistance. In addition, we also include an evaluation of several algorithms and databases for prediction and validation of circRNA and lncRNA functional interactions.
Keywords: circular RNAs; cutaneous melanoma; immunotherapy; long noncoding RNAs; targeted therapy.
© 2021 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
MM has served as a consultant and/or advisor to Roche, Bristol‐Myers Squibb, Merck Sharp Dohme, Incyte, AstraZeneca, Amgen, Pierre Fabre, Eli Lilly, Glaxo Smith Kline, SciClone, Sanofi, Alfasigma, and Merck Serono; MM and AC own shares in Epigen Therapeutics, SRL. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Noncoding RNA circuitry in melanoma onset, plasticity, and therapeutic response.Pharmacol Ther. 2023 Aug;248:108466. doi: 10.1016/j.pharmthera.2023.108466. Epub 2023 Jun 8. Pharmacol Ther. 2023. PMID: 37301330 Free PMC article. Review.
-
Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia.J Hematol Oncol. 2019 May 24;12(1):51. doi: 10.1186/s13045-019-0734-5. J Hematol Oncol. 2019. PMID: 31126316 Free PMC article. Review.
-
Interplay Between Long Noncoding RNAs and MicroRNAs in Cancer.Methods Mol Biol. 2018;1819:75-92. doi: 10.1007/978-1-4939-8618-7_4. Methods Mol Biol. 2018. PMID: 30421400
-
Circular RNAs as a Diagnostic and Therapeutic Target in Cardiovascular Diseases.Int J Mol Sci. 2023 Jan 21;24(3):2125. doi: 10.3390/ijms24032125. Int J Mol Sci. 2023. PMID: 36768449 Free PMC article. Review.
-
Mechanisms of non-coding RNA-modulated alternative splicing in cancer.RNA Biol. 2022;19(1):541-547. doi: 10.1080/15476286.2022.2062846. Epub 2021 Dec 31. RNA Biol. 2022. PMID: 35427215 Free PMC article. Review.
Cited by
-
Synergistic Immunoregulation: harnessing CircRNAs and PiRNAs to Amplify PD-1/PD-L1 Inhibition Therapy.Int J Nanomedicine. 2024 May 28;19:4803-4834. doi: 10.2147/IJN.S461289. eCollection 2024. Int J Nanomedicine. 2024. PMID: 38828205 Free PMC article. Review.
-
Non-coding RNAs in BRAF-mutant melanoma: targets, indicators, and therapeutic potential.Naunyn Schmiedebergs Arch Pharmacol. 2025 Jan;398(1):297-317. doi: 10.1007/s00210-024-03366-3. Epub 2024 Aug 21. Naunyn Schmiedebergs Arch Pharmacol. 2025. PMID: 39167168 Review.
-
circITCH suppresses cell proliferation and metastasis through miR-660/TFCP2 pathway in melanoma.Cancer Med. 2022 Jun;11(12):2405-2413. doi: 10.1002/cam4.4627. Epub 2022 Mar 10. Cancer Med. 2022. PMID: 35274492 Free PMC article.
-
Unraveling the role of hypoxia-inducible factors in cutaneous melanoma: from mechanisms to therapeutic opportunities.Cell Commun Signal. 2025 Apr 9;23(1):177. doi: 10.1186/s12964-025-02173-4. Cell Commun Signal. 2025. PMID: 40205422 Free PMC article. Review.
-
Non-Coding RNAs in Tuberculosis Epidemiology: Platforms and Approaches for Investigating the Genome's Dark Matter.Int J Mol Sci. 2022 Apr 17;23(8):4430. doi: 10.3390/ijms23084430. Int J Mol Sci. 2022. PMID: 35457250 Free PMC article. Review.
References
-
- Prado G, Svoboda RM & Rigel DS (2019) What's new in melanoma. Dermatol Clin 37, 159–168. - PubMed
-
- Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, Patch A‐M, Kakavand H, Alexandrov LB, Burke H et al. (2017) Whole‐genome landscapes of major melanoma subtypes. Nature 545, 175. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

