Identification and validation of a novel eight mutant-derived long non-coding RNAs signature as a prognostic biomarker for genome instability in low-grade glioma
- PMID: 34081618
- PMCID: PMC8221298
- DOI: 10.18632/aging.203079
Identification and validation of a novel eight mutant-derived long non-coding RNAs signature as a prognostic biomarker for genome instability in low-grade glioma
Abstract
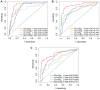
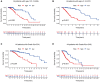
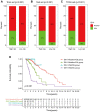
Long non-coding RNAs (lncRNAs) comprise an integral part of the eukaryotic transcriptome. Alongside proteins, lncRNAs modulate lncRNA-based gene signatures of unstable transcripts, play a crucial role as antisense lncRNAs to control intracellular homeostasis and are implicated in tumorigenesis. However, the role of genomic instability-associated lncRNAs in low-grade gliomas (LGG) has not been fully explored. In this study, lncRNAs expression and somatic mutation profiles in low-grade glioma genome were used to identify eight novel mutant-derived genomic instability-associated lncRNAs including H19, FLG-AS1, AC091932.1, AC064875.1, AL138767.3, AC010273.2, AC131097.4 and ISX-AS1. Patients from the LGG gene mutagenome atlas were grouped into training and validation sets to test the performance of the signature. The genomic instability-associated lncRNAs signature (GILncSig) was then validated using multiple external cohorts. A total of 59 novel genomic instability-associated lncRNAs in LGG were used for least absolute shrinkage and selection operator (Lasso), single and multifactor Cox regression analysis using the training set. Furthermore, the independent predictive role of risk features in the training and validation sets were evaluated through survival analysis, receiver operating feature analysis and construction of a nomogram. Patients with IDH1 mutation status were grouped into two different risk groups based on the GILncSig score. The low-risk group showed a relatively higher rate of IDH1 mutations compared with patients in the high-risk group. Furthermore, patients in the low-risk group had better prognosis compared with patients in the high-risk group. In summary, this study reports a reliable prognostic prediction signature and provides a basis for further investigation of the role of lncRNAs on genomic instability. In addition, lncRNAs in the signature can be used as new targets for treatment of LGG.
Keywords: genomic instability-associated lncRNAs signature (GILncSig); long non-coding RNA (lncRNA); low-grade glioma (LGG); prognosis; risk score.
Conflict of interest statement
Figures





Similar articles
-
Prediction of bladder cancer outcome by identifying and validating a mutation-derived genomic instability-associated long noncoding RNA (lncRNA) signature.Bioengineered. 2021 Dec;12(1):1725-1738. doi: 10.1080/21655979.2021.1924555. Bioengineered. 2021. PMID: 33955803 Free PMC article.
-
An autophagic gene-based signature to predict the survival of patients with low-grade gliomas.Cancer Med. 2021 Mar;10(5):1848-1859. doi: 10.1002/cam4.3748. Epub 2021 Feb 16. Cancer Med. 2021. PMID: 33591634 Free PMC article.
-
Identification and Validation of an Energy Metabolism-Related lncRNA-mRNA Signature for Lower-Grade Glioma.Biomed Res Int. 2020 Jul 27;2020:3708231. doi: 10.1155/2020/3708231. eCollection 2020. Biomed Res Int. 2020. PMID: 32802843 Free PMC article.
-
Long non-coding RNAs in glioma progression.Cancer Lett. 2018 Apr 10;419:203-209. doi: 10.1016/j.canlet.2018.01.041. Epub 2018 Jan 31. Cancer Lett. 2018. PMID: 29355660 Review.
-
Radiomic Prediction of CCND1 Expression Levels and Prognosis in Low-grade Glioma Based on Magnetic Resonance Imaging.Acad Radiol. 2024 Nov;31(11):4595-4610. doi: 10.1016/j.acra.2024.03.031. Epub 2024 Jun 1. Acad Radiol. 2024. PMID: 38824087 Review.
Cited by
-
Multi-Omics Analysis Based on Genomic Instability for Prognostic Prediction in Lower-Grade Glioma.Front Genet. 2022 Jan 5;12:758596. doi: 10.3389/fgene.2021.758596. eCollection 2021. Front Genet. 2022. PMID: 35069679 Free PMC article.
-
Immune-relatedlncRNAs can predict the prognosis of acute myeloid leukemia.Cancer Med. 2022 Feb;11(3):888-899. doi: 10.1002/cam4.4487. Epub 2021 Dec 14. Cancer Med. 2022. PMID: 34904791 Free PMC article.
-
TMEM71 is crucial for cell proliferation in lower-grade glioma and is linked to unfavorable prognosis.Cancer Cell Int. 2025 Mar 21;25(1):109. doi: 10.1186/s12935-025-03747-5. Cancer Cell Int. 2025. PMID: 40119335 Free PMC article.
-
Single cell RNA sequencing reveals differentiation related genes with drawing implications in predicting prognosis and immunotherapy response in gliomas.Sci Rep. 2022 Feb 3;12(1):1872. doi: 10.1038/s41598-022-05686-x. Sci Rep. 2022. PMID: 35115572 Free PMC article.
-
Novel GIRlncRNA Signature for Predicting the Clinical Outcome and Therapeutic Response in NSCLC.Front Pharmacol. 2022 Aug 3;13:937531. doi: 10.3389/fphar.2022.937531. eCollection 2022. Front Pharmacol. 2022. PMID: 35991889 Free PMC article.
References
-
- Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016; 131:803–20. 10.1007/s00401-016-1545-1 - DOI - PubMed
-
- Lu VM, O'Connor KP, Shah AH, Eichberg DG, Luther EM, Komotar RJ, Ivan ME. The prognostic significance of CDKN2A homozygous deletion in IDH-mutant lower-grade glioma and glioblastoma: a systematic review of the contemporary literature. J Neurooncol. 2020; 148:221–29. 10.1007/s11060-020-03528-2 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

