GCSENet: A GCN, CNN and SENet ensemble model for microRNA-disease association prediction
- PMID: 34081706
- PMCID: PMC8205154
- DOI: 10.1371/journal.pcbi.1009048
GCSENet: A GCN, CNN and SENet ensemble model for microRNA-disease association prediction
Abstract
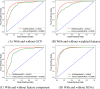
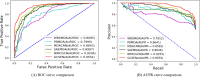
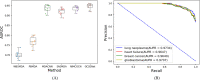
Recently, an increasing number of studies have demonstrated that miRNAs are involved in human diseases, indicating that miRNAs might be a potential pathogenic factor for various diseases. Therefore, figuring out the relationship between miRNAs and diseases plays a critical role in not only the development of new drugs, but also the formulation of individualized diagnosis and treatment. As the prediction of miRNA-disease association via biological experiments is expensive and time-consuming, computational methods have a positive effect on revealing the association. In this study, a novel prediction model integrating GCN, CNN and Squeeze-and-Excitation Networks (GCSENet) was constructed for the identification of miRNA-disease association. The model first captured features by GCN based on a heterogeneous graph including diseases, genes and miRNAs. Then, considering the different effects of genes on each type of miRNA and disease, as well as the different effects of the miRNA-gene and disease-gene relationships on miRNA-disease association, a feature weight was set and a combination of miRNA-gene and disease-gene associations was added as feature input for the convolution operation in CNN. Furthermore, the squeeze and excitation blocks of SENet were applied to determine the importance of each feature channel and enhance useful features by means of the attention mechanism, thus achieving a satisfactory prediction of miRNA-disease association. The proposed method was compared against other state-of-the-art methods. It achieved an AUROC score of 95.02% and an AUPR score of 95.55% in a 10-fold cross-validation, which led to the finding that the proposed method is superior to these popular methods on most of the performance evaluation indexes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


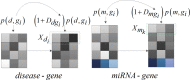


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

