Robustness of epithelial sealing is an emerging property of local ERK feedback driven by cell elimination
- PMID: 34081909
- PMCID: PMC8221813
- DOI: 10.1016/j.devcel.2021.05.006
Robustness of epithelial sealing is an emerging property of local ERK feedback driven by cell elimination
Abstract
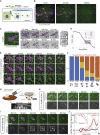
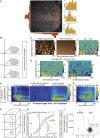
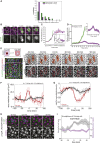
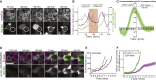
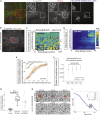
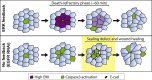
What regulates the spatiotemporal distribution of cell elimination in tissues remains largely unknown. This is particularly relevant for epithelia with high rates of cell elimination where simultaneous death of neighboring cells could impair epithelial sealing. Here, using the Drosophila pupal notum (a single-layer epithelium) and a new optogenetic tool to trigger caspase activation and cell extrusion, we first showed that death of clusters of at least three cells impaired epithelial sealing; yet, such clusters were almost never observed in vivo. Accordingly, statistical analysis and simulations of cell death distribution highlighted a transient and local protective phase occurring near every cell death. This protection is driven by a transient activation of ERK in cells neighboring extruding cells, which inhibits caspase activation and prevents elimination of cells in clusters. This suggests that the robustness of epithelia with high rates of cell elimination is an emerging property of local ERK feedback.
Keywords: Drosophila; ERK; apoptosis; caspase dynamics; cell extrusion; epithelium; optogenetics; self-organization; single-cell live imaging.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
-
- Aigouy B., Farhadifar R., Staple D.B., Sagner A., Röper J.C., Jülicher F., Eaton S. Cell flow reorients the axis of planar polarity in the wing epithelium of Drosophila. Cell. 2010;142:773–786. - PubMed
-
- Aoki K., Kondo Y., Naoki H., Hiratsuka T., Itoh R.E., Matsuda M. Propagating wave of ERK activation orients collective cell migration. Dev. Cell. 2017;43:305–317.e5. - PubMed
-
- Aoki K., Kumagai Y., Sakurai A., Komatsu N., Fujita Y., Shionyu C., Matsuda M. Stochastic ERK activation induced by noise and cell-to-cell propagation regulates cell density-dependent proliferation. Mol. Cell. 2013;52:529–540. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

