SARS-CoV-2 infection induces beta cell transdifferentiation
- PMID: 34081913
- PMCID: PMC8133495
- DOI: 10.1016/j.cmet.2021.05.015
SARS-CoV-2 infection induces beta cell transdifferentiation
Abstract
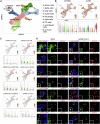
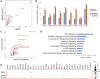
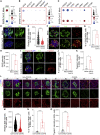
Recent clinical data have suggested a correlation between coronavirus disease 2019 (COVID-19) and diabetes. Here, we describe the detection of SARS-CoV-2 viral antigen in pancreatic beta cells in autopsy samples from individuals with COVID-19. Single-cell RNA sequencing and immunostaining from ex vivo infections confirmed that multiple types of pancreatic islet cells were susceptible to SARS-CoV-2, eliciting a cellular stress response and the induction of chemokines. Upon SARS-CoV-2 infection, beta cells showed a lower expression of insulin and a higher expression of alpha and acinar cell markers, including glucagon and trypsin1, respectively, suggesting cellular transdifferentiation. Trajectory analysis indicated that SARS-CoV-2 induced eIF2-pathway-mediated beta cell transdifferentiation, a phenotype that could be reversed with trans-integrated stress response inhibitor (trans-ISRIB). Altogether, this study demonstrates an example of SARS-CoV-2 infection causing cell fate change, which provides further insight into the pathomechanisms of COVID-19.
Keywords: COVID-19; EgIF2; PRSS1; diabetes; human islets; insulin; trypsin 1.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.E.S. is on the scientific advisory board of Miromatrix Inc. and is a paid consultant and speaker for Alnylam Inc. The other authors declare no competing interests.
Figures





Comment in
-
SARS-CoV-2 infection of islet β cells: Evidence and implications.Cell Rep Med. 2021 Aug 17;2(8):100380. doi: 10.1016/j.xcrm.2021.100380. Cell Rep Med. 2021. PMID: 34423322 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- DP3 DK111907/DK/NIDDK NIH HHS/United States
- R01 DK119667/DK/NIDDK NIH HHS/United States
- R01 CA234614/CA/NCI NIH HHS/United States
- ZIA HG000024/ImNIH/Intramural NIH HHS/United States
- R01 DK124463/DK/NIDDK NIH HHS/United States
- R03 DK117252/DK/NIDDK NIH HHS/United States
- R01 DK116075/DK/NIDDK NIH HHS/United States
- UC4 DK112217/DK/NIDDK NIH HHS/United States
- R01 AI107301/AI/NIAID NIH HHS/United States
- F32 HD096810/HD/NICHD NIH HHS/United States
- U01 DK127777/DK/NIDDK NIH HHS/United States
- P30 DK020541/DK/NIDDK NIH HHS/United States
- R01 DK121072/DK/NIDDK NIH HHS/United States
- P60 DK020541/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

