The case for AI-driven cancer clinical trials - The efficacy arm in silico
- PMID: 34082064
- PMCID: PMC8922906
- DOI: 10.1016/j.bbcan.2021.188572
The case for AI-driven cancer clinical trials - The efficacy arm in silico
Abstract
Pharmaceutical agents in oncology currently have high attrition rates from early to late phase clinical trials. Recent advances in computational methods, notably causal artificial intelligence, and availability of rich clinico-genomic databases have made it possible to simulate the efficacy of cancer drug protocols in diverse patient populations, which could inform and improve clinical trial design. Here, we review the current and potential use of in silico trials and causal AI to increase the efficacy and safety of traditional clinical trials. We conclude that in silico trials using causal AI approaches can simulate control and efficacy arms, inform patient recruitment and regimen titrations, and better enable subgroup analyses critical for precision medicine.
Keywords: AI; Causal AI; Disease modeling; Efficacy arm; Systems biomedicine; in silico clinical trials.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest
The authors declare that they have the following competing financial interests or personal relationships: RBP has received personal fees and equity from GNS Healthcare. FG and CH are employees of GNS Healthcare.
Figures
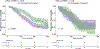
References
-
- Auslander N, Zhang G, Lee JS, Frederick DT, Miao B, Moll T, Tian T, Wei Z, Madan S, Sullivan RJ, Boland G, Flaherty K, Herlyn M, & Ruppin E (2018). Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nature Medicine, 24(10), 1545–1549. doi:10.1038/s41591-018-0157-9 - DOI - PMC - PubMed
-
- Badano A, Graff CG, Badal A, Sharma D, Zeng R, Samuelson FW, Glick SJ, & Myers KJ (2018). Evaluation of Digital Breast Tomosynthesis as Replacement of Full-Field Digital Mammography Using an In Silico Imaging Trial. JAMA Network Open, 1(7), e185474. doi:10.1001/jamanetworkopen.2018.5474 - DOI - PMC - PubMed
-
- Bianchi A (2013). Patient Recruitment Driving Length and Cost of Oncology Clinical Trials. International Pharmaceutical Industry, 5(2), 58–61. http://ipimediaworld.com/wp-content/uploads/2013/06/2-Patient-Recruitmen...“ http://ipimediaworld.com/wp-content/uploads/2013/06/2-Patient-Recruitmen...
-
- Bolomsky A, Gruber F, Stangelberger K, Furchtgott L, Arnold D, Raut P, Wuest D, Runge K, Khalil I, Zojer N, Munshi N, Hayete B, Ludwig H (2018). Preclinical Validation Studies Support Causal Machine Learning Based Identification of Novel Drug Targets for High-Risk Multiple Myeloma. Blood 132, 3210–3210. doi:10.1182/blood-2018-99-117886 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

