Mice with dysfunctional TGF-β signaling develop altered intestinal microbiome and colorectal cancer resistant to 5FU
- PMID: 34082069
- PMCID: PMC8815765
- DOI: 10.1016/j.bbadis.2021.166179
Mice with dysfunctional TGF-β signaling develop altered intestinal microbiome and colorectal cancer resistant to 5FU
Abstract
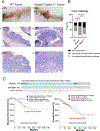
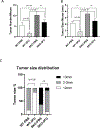
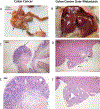
Emerging data show a rise in colorectal cancer (CRC) incidence in young men and women that is often chemoresistant. One potential risk factor is an alteration in the microbiome. Here, we investigated the role of TGF-β signaling on the intestinal microbiome and the efficacy of chemotherapy for CRC induced by azoxymethane and dextran sodium sulfate in mice. We used two genotypes of TGF-β-signaling-deficient mice (Smad4+/- and Smad4+/-Sptbn1+/-), which developed CRC with similar phenotypes and had similar alterations in the intestinal microbiome. Using these mice, we evaluated the intestinal microbiome and determined the effect of dysfunctional TGF-β signaling on the response to the chemotherapeutic agent 5-Fluoro-uracil (5FU) after induction of CRC. Using shotgun metagenomic sequencing, we determined gut microbiota composition in mice with CRC and found reduced amounts of beneficial species of Bacteroides and Parabacteroides in the mutants compared to the wild-type (WT) mice. Furthermore, the mutant mice with CRC were resistant to 5FU. Whereas the abundances of E. boltae, B.dorei, Lachnoclostridium sp., and Mordavella sp. were significantly reduced in mice with CRC, these species only recovered to basal amounts after 5FU treatment in WT mice, suggesting that the alterations in the intestinal microbiome resulting from compromised TGF-β signaling impaired the response to 5FU. These findings could have implications for inhibiting the TGF-β pathway in the treatment of CRC or other cancers.
Keywords: 5FU; Bacteroides; Chemoresistance; Colorectal cancer; Microbiome; TGF-β signaling.
Copyright © 2021. Published by Elsevier B.V.
Conflict of interest statement
Figures





References
-
- Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A, Colorectal cancer statistics, 2020, CA Cancer J Clin 70(3) (2020) 145–164. - PubMed
-
- Blaj C, Schmidt EM, Lamprecht S, Hermeking H, Jung A, Kirchner T, Horst D, Oncogenic Effects of High MAPK Activity in Colorectal Cancer Mark Progenitor Cells and Persist Irrespective of RAS Mutations, Cancer Res 77(7) (2017) 1763–1774. - PubMed
-
- Mishra L, Shetty K, Tang Y, Stuart A, Byers SW, The role of TGF-beta and Wnt signaling in gastrointestinal stem cells and cancer, Oncogene 24(37) (2005) 5775–89. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

