Post-diapause transcriptomic restarts: insight from a high-latitude copepod
- PMID: 34082716
- PMCID: PMC8176732
- DOI: 10.1186/s12864-021-07557-7
Post-diapause transcriptomic restarts: insight from a high-latitude copepod
Erratum in
-
Correction to: Post-diapause transcriptomic restarts: insight from a high-latitude copepod.BMC Genomics. 2021 Sep 14;22(1):659. doi: 10.1186/s12864-021-07868-9. BMC Genomics. 2021. PMID: 34521338 Free PMC article. No abstract available.
Abstract
Background: Diapause is a seasonal dormancy that allows organisms to survive unfavorable conditions and optimizes the timing of reproduction and growth. Emergence from diapause reverses the state of arrested development and metabolic suppression returning the organism to an active state. The physiological mechanisms that regulate the transition from diapause to post-diapause are still unknown. In this study, this transition has been characterized for the sub-arctic calanoid copepod Neocalanus flemingeri, a key crustacean zooplankter that supports the highly productive North Pacific fisheries. Transcriptional profiling of females, determined over a two-week time series starting with diapausing females collected from > 400 m depth, characterized the molecular mechanisms that regulate the post-diapause trajectory.
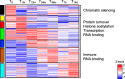
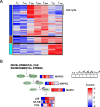
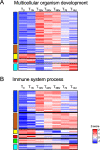
Results: A complex set of transitions in relative gene expression defined the transcriptomic changes from diapause to post-diapause. Despite low temperatures (5-6 °C), the switch from a "diapause" to a "post-diapause" transcriptional profile occurred within 12 h of the termination stimulus. Transcriptional changes signaling the end of diapause were activated within one-hour post collection and included the up-regulation of genes involved in the 20E cascade pathway, the TCA cycle and RNA metabolism in combination with the down-regulation of genes associated with chromatin silencing. By 12 h, females exhibited a post-diapause phenotype characterized by the up-regulation of genes involved in cell division, cell differentiation and multiple developmental processes. By seven days post collection, the reproductive program was fully activated as indicated by up-regulation of genes involved in oogenesis and energy metabolism, processes that were enriched among the differentially expressed genes.
Conclusions: The analysis revealed a finely structured, precisely orchestrated sequence of transcriptional changes that led to rapid changes in the activation of biological processes paving the way to the successful completion of the reproductive program. Our findings lead to new hypotheses related to potentially universal mechanisms that terminate diapause before an organism can resume its developmental program.
Keywords: Copepod; Diapause; Gulf of Alaska; Neocalanus flemingeri; Respiration; Transcriptomics.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

