The global gene expression outline of the bovine blastocyst: reflector of environmental conditions and predictor of developmental capacity
- PMID: 34082721
- PMCID: PMC8176733
- DOI: 10.1186/s12864-021-07693-0
The global gene expression outline of the bovine blastocyst: reflector of environmental conditions and predictor of developmental capacity
Abstract
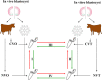
Background: Morphological evaluation of embryos has been used to screen embryos for transfer. However, the repeatability and accuracy of this method remains low. Thus, evaluation of an embryo's gene expression signature with respect to its developmental capacity could provide new opportunities for embryo selection. Since the gene expression outline of an embryo is considered as an aggregate of its intrinsic characteristics and culture conditions, we have compared transcriptome profiles of in vivo and in vitro derived blastocysts in relation to pregnancy outcome to unravel the discrete effects of developmental competence and environmental conditions on bovine embryo gene expression outlines. To understand whether the gene expression patterns could be associated with blastocyst developmental competency, the global transcriptome profile of in vivo (CVO) and in vitro (CVT) derived competent blastocysts that resulted in pregnancy was investigated relative to that of in vivo (NVO) and in vitro (NVT) derived blastocysts which did not establish initial pregnancy, respectively while to unravel the effects of culture condition on the transcriptome profile of embryos, the transcriptional activity of the CVO group was compared to the CVT group and the NVO group was compared to the NVT ones.
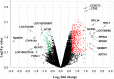
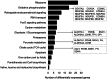
Results: A total of 700 differentially expressed genes (DEGs) were identified between CVO and NVO blastocysts. These gene transcripts represent constitutive regions, indel variants, 3'-UTR sequence variants and novel transcript regions. The majority (82%) of these DEGs, including gene clusters like ATP synthases, eukaryotic translation initiation factors, ribosomal proteins, mitochondrial ribosomal proteins, NADH dehydrogenase and cytochrome c oxidase subunits were enriched in the CVO group. These DEGs were involved in pathways associated with glycolysis/glycogenesis, citrate acid cycle, pyruvate metabolism and oxidative phosphorylation. Similarly, a total of 218 genes were differentially expressed between CVT and NVT groups. Of these, 89%, including TPT1, PDIA6, HSP90AA1 and CALM, were downregulated in the CVT group and those DEGs were overrepresented in pathways related to protein processing, endoplasmic reticulum, spliceasome, ubiquitone mediated proteolysis and steroid biosynthesis. On the other hand, although both the CVT and CVO blastocyst groups resulted in pregnancy, a total of 937 genes were differential expressed between the two groups. Compared to CVO embryos, the CVT ones exhibited downregulation of gene clusters including ribosomal proteins, mitochondrial ribosomal protein, eukaryotic translation initiation factors, ATP synthases, NADH dehydrogenase and cytochrome c oxidases. Nonetheless, downregulation of these genes could be associated with pre and postnatal abnormalities observed after transfer of in vitro embryos.
Conclusion: The present study provides a detailed inventory of differentially expressed gene signatures and pathways specifically reflective of the developmental environment and future developmental capacities of bovine embryos suggesting that transcriptome activity observed in blastocysts could be indicative of further pregnancy success but also adaptation to culture environment.
Keywords: Bovine; Embryo; Pregnancy; Transcriptome.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures










Similar articles
-
Deregulation of oxidative phosphorylation pathways in embryos derived in vitro from prepubertal and pubertal heifers based on whole-transcriptome sequencing.BMC Genomics. 2024 Jun 24;25(1):632. doi: 10.1186/s12864-024-10532-7. BMC Genomics. 2024. PMID: 38914933 Free PMC article.
-
Bovine blastocysts with developmental competence to term share similar expression of developmentally important genes although derived from different culture environments.Reproduction. 2011 Oct;142(4):551-64. doi: 10.1530/REP-10-0476. Epub 2011 Jul 28. Reproduction. 2011. PMID: 21799070
-
Genome-Wide DNA Methylation Patterns of Bovine Blastocysts Developed In Vivo from Embryos Completed Different Stages of Development In Vitro.PLoS One. 2015 Nov 4;10(11):e0140467. doi: 10.1371/journal.pone.0140467. eCollection 2015. PLoS One. 2015. PMID: 26536655 Free PMC article.
-
Review: Overview of the transcriptomic landscape in bovine blastocysts and elongated conceptuses driving developmental competence.Animal. 2023 May;17 Suppl 1:100733. doi: 10.1016/j.animal.2023.100733. Animal. 2023. PMID: 37567651 Review.
-
Gene expression analysis and in vitro production procedures for bovine preimplantation embryos: Past highlights, present concepts and future prospects.Reprod Domest Anim. 2018 Sep;53 Suppl 2:14-19. doi: 10.1111/rda.13260. Reprod Domest Anim. 2018. PMID: 30238652 Review.
Cited by
-
Lipid Enriched Reduced Nutrient Culture Medium Improves Bovine Blastocyst Formation.Reprod Fertil. 2023 Nov 1;4(4):e230057. doi: 10.1530/RAF-23-0057. Online ahead of print. Reprod Fertil. 2023. PMID: 37971749 Free PMC article.
-
Development of a formula for scoring competence of bovine embryos to sustain pregnancy.Biochem Biophys Rep. 2024 Jul 2;39:101772. doi: 10.1016/j.bbrep.2024.101772. eCollection 2024 Sep. Biochem Biophys Rep. 2024. PMID: 39050012 Free PMC article.
-
The mitochondrial respiration signature of the bovine blastocyst reflects both environmental conditions of development as well as embryo quality.Sci Rep. 2023 Nov 8;13(1):19408. doi: 10.1038/s41598-023-45691-2. Sci Rep. 2023. PMID: 37938581 Free PMC article.
-
Proteomic Analysis of Domestic Cat Blastocysts and Their Secretome Produced in an In Vitro Culture System without the Presence of the Zona Pellucida.Int J Mol Sci. 2024 Apr 14;25(8):4343. doi: 10.3390/ijms25084343. Int J Mol Sci. 2024. PMID: 38673927 Free PMC article.
-
Sexual dimorphic miRNA-mediated response of bovine elongated embryos to the maternal microenvironment.PLoS One. 2024 Feb 29;19(2):e0298835. doi: 10.1371/journal.pone.0298835. eCollection 2024. PLoS One. 2024. PMID: 38422042 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

