Atg7 deficiency in microglia drives an altered transcriptomic profile associated with an impaired neuroinflammatory response
- PMID: 34082793
- PMCID: PMC8173846
- DOI: 10.1186/s13041-021-00794-7
Atg7 deficiency in microglia drives an altered transcriptomic profile associated with an impaired neuroinflammatory response
Abstract
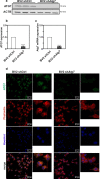
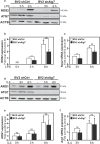
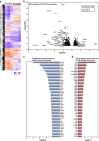
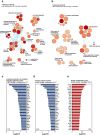
Microglia, resident immunocompetent cells of the central nervous system, can display a range of reaction states and thereby exhibit distinct biological functions across development, adulthood and under disease conditions. Distinct gene expression profiles are reported to define each of these microglial reaction states. Hence, the identification of modulators of selective microglial transcriptomic signature, which have the potential to regulate unique microglial function has gained interest. Here, we report the identification of ATG7 (Autophagy-related 7) as a selective modulator of an NF-κB-dependent transcriptional program controlling the pro-inflammatory response of microglia. We also uncover that microglial Atg7-deficiency was associated with reduced microglia-mediated neurotoxicity, and thus a loss of biological function associated with the pro-inflammatory microglial reactive state. Further, we show that Atg7-deficiency in microglia did not impact on their ability to respond to alternative stimulus, such as one driving them towards an anti-inflammatory/tumor supportive phenotype. The identification of distinct regulators, such as Atg7, controlling specific microglial transcriptional programs could lead to developing novel therapeutic strategies aiming to manipulate selected microglial phenotypes, instead of the whole microglial population with is associated with several pitfalls.
Keywords: Atg7; Inflammatory response; Microglia; NF-κB; Neurotoxicity; Transcriptome.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
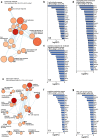


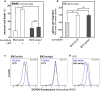
Similar articles
-
DJ-1 inhibits microglial activation and protects dopaminergic neurons in vitro and in vivo through interacting with microglial p65.Cell Death Dis. 2021 Jul 17;12(8):715. doi: 10.1038/s41419-021-04002-1. Cell Death Dis. 2021. PMID: 34274951 Free PMC article.
-
Mineralocorticoid and glucocorticoid receptors differentially regulate NF-kappaB activity and pro-inflammatory cytokine production in murine BV-2 microglial cells.J Neuroinflammation. 2012 Nov 28;9:260. doi: 10.1186/1742-2094-9-260. J Neuroinflammation. 2012. PMID: 23190711 Free PMC article.
-
Tripartite-motif protein 21 knockdown extenuates LPS-triggered neurotoxicity by inhibiting microglial M1 polarization via suppressing NF-κB-mediated NLRP3 inflammasome activation.Arch Biochem Biophys. 2021 Jul 30;706:108918. doi: 10.1016/j.abb.2021.108918. Epub 2021 May 13. Arch Biochem Biophys. 2021. PMID: 33992596
-
Epigenetic control of microglial immune responses.Immunol Rev. 2024 May;323(1):209-226. doi: 10.1111/imr.13317. Epub 2024 Mar 16. Immunol Rev. 2024. PMID: 38491845 Review.
-
Microglial subtypes: diversity within the microglial community.EMBO J. 2019 Sep 2;38(17):e101997. doi: 10.15252/embj.2019101997. Epub 2019 Aug 2. EMBO J. 2019. PMID: 31373067 Free PMC article. Review.
Cited by
-
Studying Autophagy in Microglia: Overcoming the Obstacles.Methods Mol Biol. 2024;2713:45-70. doi: 10.1007/978-1-0716-3437-0_3. Methods Mol Biol. 2024. PMID: 37639114
-
Intervention of NF-Κb Signaling Pathway and Preventing Post-Operative Cognitive Dysfunction as Well as Neuronal Apoptosis.Iran J Public Health. 2022 Jan;51(1):124-132. doi: 10.18502/ijph.v51i1.8303. Iran J Public Health. 2022. PMID: 35223633 Free PMC article.
-
Endothelial autophagy-related gene 7 contributes to high fat diet-induced obesity.Mol Metab. 2025 Mar;93:102099. doi: 10.1016/j.molmet.2025.102099. Epub 2025 Jan 18. Mol Metab. 2025. PMID: 39832563 Free PMC article.
-
Autophagy in alzheimer disease pathogenesis and its therapeutic values.Autophagy Rep. 2025 May 8;4(1):2471677. doi: 10.1080/27694127.2025.2471677. eCollection 2025. Autophagy Rep. 2025. PMID: 40395993 Free PMC article. Review.
-
Uncovering ferroptosis in Parkinson's disease via bioinformatics and machine learning, and reversed deducing potential therapeutic natural products.Front Genet. 2023 Jul 6;14:1231707. doi: 10.3389/fgene.2023.1231707. eCollection 2023. Front Genet. 2023. PMID: 37485340 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

