Genomic and transcriptomic analysis of the thermophilic lignocellulose-degrading fungus Thielavia terrestris LPH172
- PMID: 34082802
- PMCID: PMC8176577
- DOI: 10.1186/s13068-021-01975-1
Genomic and transcriptomic analysis of the thermophilic lignocellulose-degrading fungus Thielavia terrestris LPH172
Abstract
Background: Biomass-degrading enzymes with improved activity and stability can increase substrate saccharification and make biorefineries economically feasible. Filamentous fungi are a rich source of carbohydrate-active enzymes (CAZymes) for biomass degradation. The newly isolated LPH172 strain of the thermophilic Ascomycete Thielavia terrestris has been shown to possess high xylanase and cellulase activities and tolerate low pH and high temperatures. Here, we aimed to illuminate the lignocellulose-degrading machinery and novel carbohydrate-active enzymes in LPH172 in detail.
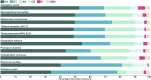
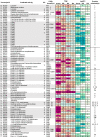
Results: We sequenced and analyzed the 36.6-Mb genome and transcriptome of LPH172 during growth on glucose, cellulose, rice straw, and beechwood xylan. 10,128 predicted genes were found in total, which included 411 CAZy domains. Compared to other fungi, auxiliary activity (AA) domains were particularly enriched. A higher GC content was found in coding sequences compared to the overall genome, as well as a high GC3 content, which is hypothesized to contribute to thermophilicity. Primarily auxiliary activity (AA) family 9 lytic polysaccharide monooxygenase (LPMO) and glycoside hydrolase (GH) family 7 glucanase encoding genes were upregulated when LPH172 was cultivated on cellulosic substrates. Conventional hemicellulose encoding genes (GH10, GH11 and various CEs), as well as AA9 LPMOs, were upregulated when LPH172 was cultivated on xylan. The observed co-expression and co-upregulation of genes encoding AA9 LPMOs, other AA CAZymes, and (hemi)cellulases point to a complex and nuanced degradation strategy.
Conclusions: Our analysis of the genome and transcriptome of T. terrestris LPH172 elucidates the enzyme arsenal that the fungus uses to degrade lignocellulosic substrates. The study provides the basis for future characterization of potential new enzymes for industrial biomass saccharification.
Keywords: Biomass degradation; Carbohydrate active enzymes; Cellulose; Filamentous fungi; LPMO; Thermostable enzymes; Transcriptome; Xylan.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

