Exosomal circ-BRWD1 contributes to osteoarthritis development through the modulation of miR-1277/TRAF6 axis
- PMID: 34082824
- PMCID: PMC8173917
- DOI: 10.1186/s13075-021-02541-8
Exosomal circ-BRWD1 contributes to osteoarthritis development through the modulation of miR-1277/TRAF6 axis
Abstract
Background: Circular RNAs (circRNAs) can act as vital players in osteoarthritis (OA). However, the roles of circRNAs in OA remain obscure. Herein, we explored the roles of exosomal circRNA bromodomain and WD repeat domain containing 1(circ-BRWD1) in OA pathology.
Methods: In vitro model of OA was constructed by treating CHON-001 cells with interleukin-1β (IL-1β). Quantitative real-time polymerase chain reaction (qRT-PCR) assay was used for circ-BRWD1, BRWD, miR-1277, and TNF receptor-associated factor 6 (TRAF6) levels. RNase R assay was conducted for the feature of circ-BRWD1. Transmission electron microscopy (TEM) was employed to analyze the morphology of exosomes. Western blot assay was performed for protein levels. Cell Counting Kit-8 (CCK-8) assay, flow cytometry analysis, and 5-Ethynyl-2'-deoxyuridine (EDU) assay were adopted for cell viability, apoptosis, and proliferation, respectively. Enzyme-linked immunosorbent assay (ELISA) was carried out for the concentrations of interleukin-6 (IL-6) and interleukin-8 (IL-8). Dual-luciferase reporter and RNA immunoprecipitation (RIP) assays were used to analyze the interaction between miR-1277 and circ-BRWD1 or TRAF6.
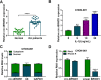
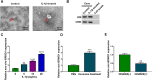
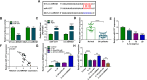
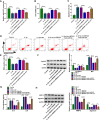
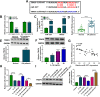
Results: Circ-BRWD1 was increased in OA cartilage tissues, IL-1β-treated CHON-001 cells, and the exosomes derived from IL-1β-treated CHON-001 cells. Exosome treatment elevated circ-BRWD1 level, while exosome blocker reduced circ-BRWD1 level in IL-1β-treated CHON-001 cells. Silencing of circ-BRWD1 promoted cell viability and proliferation and repressed apoptosis, inflammation, and extracellular matrix (ECM) degradation in IL-1β-stimulated CHON-001 cells. For mechanism analysis, circ-BRWD1 could serve as the sponge for miR-1277 to positively regulate TRAF6 expression. Moreover, miR-1277 inhibition ameliorated the effects of circ-BRWD1 knockdown on IL-1β-mediated CHON-001 cell damage. Additionally, miR-1277 overexpression relieved IL-1β-induced CHON-001 cell injury, while TRAF6 elevation restored the impact.
Conclusion: Exosomal circ-BRWD1 promoted IL-1β-induced CHON-001 cell progression by regulating miR-1277/TRAF6 axis.
Keywords: Circ-BRWD1; Exosome; MiR-1277; OA; TRAF6.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
CircRNA circ-IQGAP1 Knockdown Alleviates Interleukin-1β-Induced Osteoarthritis Progression via Targeting miR-671-5p/TCF4.Orthop Surg. 2021 May;13(3):1036-1046. doi: 10.1111/os.12923. Epub 2021 Mar 5. Orthop Surg. 2021. PMID: 33675175 Free PMC article.
-
Circular RNA circ_0008365 regulates SOX9 by targeting miR-338-3p to inhibit IL-1β-induced chondrocyte apoptosis and extracellular matrix degradation.J Orthop Surg Res. 2022 Oct 14;17(1):452. doi: 10.1186/s13018-022-03240-z. J Orthop Surg Res. 2022. PMID: 36242088 Free PMC article.
-
Knockdown of circ-PRKCH alleviates IL-1β-treated chondrocyte cell phenotypic changes through modulating miR-502-5p/ADAMTS5 axis.Autoimmunity. 2022 May;55(3):179-191. doi: 10.1080/08916934.2022.2027918. Epub 2022 Mar 30. Autoimmunity. 2022. PMID: 35352613
-
CircRNAs in osteoarthritis: research status and prospect.Front Genet. 2023 May 9;14:1173812. doi: 10.3389/fgene.2023.1173812. eCollection 2023. Front Genet. 2023. PMID: 37229197 Free PMC article. Review.
-
Regulatory mechanism of circular RNA involvement in osteoarthritis.Front Surg. 2023 Jan 6;9:1049513. doi: 10.3389/fsurg.2022.1049513. eCollection 2022. Front Surg. 2023. PMID: 36684373 Free PMC article. Review.
Cited by
-
The Role of Extracellular Vesicles in the Pathogenesis and Treatment of Rheumatoid Arthritis and Osteoarthritis.Cells. 2023 Nov 27;12(23):2716. doi: 10.3390/cells12232716. Cells. 2023. PMID: 38067147 Free PMC article. Review.
-
The communication role of extracellular vesicles in the osteoarthritis microenvironment.Front Immunol. 2025 Mar 17;16:1549833. doi: 10.3389/fimmu.2025.1549833. eCollection 2025. Front Immunol. 2025. PMID: 40165965 Free PMC article. Review.
-
Circular RNA triple functional domain promotes osteoarthritis' development by modulating the microRNA-136-5p/Nicotinamide phosphoribosyltransferase axis.Bioengineered. 2022 Mar;13(3):6070-6079. doi: 10.1080/21655979.2021.2018095. Bioengineered. 2022. PMID: 35191807 Free PMC article.
-
Extracellular vesicles from the inflammatory microenvironment regulate the osteogenic and odontogenic differentiation of periodontal ligament stem cells by miR-758-5p/LMBR1/BMP2/4 axis.J Transl Med. 2022 May 13;20(1):208. doi: 10.1186/s12967-022-03412-9. J Transl Med. 2022. PMID: 35562763 Free PMC article.
-
Role of Exosomal Non-Coding RNAs in Bone-Related Diseases.Front Cell Dev Biol. 2021 Dec 23;9:811666. doi: 10.3389/fcell.2021.811666. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 35004702 Free PMC article. Review.
References
-
- Honvo G, Reginster JY, Rabenda V, Geerinck A, Mkinsi O, Charles A, Rizzoli R, Cooper C, Avouac B, Bruyère O. Safety of symptomatic slow-acting drugs for osteoarthritis: outcomes of a systematic review and meta-analysis. Drugs Aging. 2019;36(Suppl 1):65–99. doi: 10.1007/s40266-019-00662-z. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

