Function and molecular mechanisms of APE2 in genome and epigenome integrity
- PMID: 34083046
- PMCID: PMC8287789
- DOI: 10.1016/j.mrrev.2020.108347
Function and molecular mechanisms of APE2 in genome and epigenome integrity
Abstract
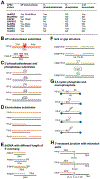
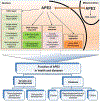
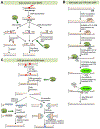
APE2 is a rising vital player in the maintenance of genome and epigenome integrity. In the past several years, a series of studies have shown the critical roles and functions of APE2. We seek to provide the first comprehensive review on several aspects of APE2 in genome and epigenome integrity. We first summarize the distinct functional domains or motifs within APE2 including EEP (endonuclease/exonuclease/phosphatase) domain, PIP box and Zf-GRF motifs from eight species (i.e., Homo sapiens, Mus musculus, Xenopus laevis, Ciona intestinalis, Arabidopsis thaliana, Schizosaccharomyces pombe, Saccharomyces cerevisiae, and Trypanosoma cruzi). Then we analyze various APE2 nuclease activities and associated DNA substrates, including AP endonuclease, 3'-phosphodiesterase, 3'-phosphatase, and 3'-5' exonuclease activities. We also examine several APE2 interaction proteins, including PCNA, Chk1, APE1, Myh1, and homologous recombination (HR) factors such as Rad51, Rad52, BRCA1, BRCA2, and BARD1. Furthermore, we provide insights into the roles of APE2 in various DNA repair pathways (base excision repair, single-strand break repair, and double-strand break repair), DNA damage response (DDR) pathways (ATR-Chk1 and p53-dependent), immunoglobulin class switch recombination and somatic hypermutation, as well as active DNA demethylation. Lastly, we summarize critical functions of APE2 in growth, development, and diseases. In this review, we provide the first comprehensive perspective which dissects all aspects of the multiple-function protein APE2 in genome and epigenome integrity.
Keywords: APE2; ATR-Chk1 pathway; DNA demethylation; DNA repair; Genome and epigenome integrity; Immune response.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors report no declarations of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

