Modeling Clinical Responses to Targeted Therapies by Patient-Derived Organoids of Advanced Lung Adenocarcinoma
- PMID: 34083237
- PMCID: PMC9401503
- DOI: 10.1158/1078-0432.CCR-20-5026
Modeling Clinical Responses to Targeted Therapies by Patient-Derived Organoids of Advanced Lung Adenocarcinoma
Abstract
Purpose: Patient-derived organoids (PDO) of lung cancer has been recently introduced, reflecting the genomic landscape of lung cancer. However, clinical relevance of advanced lung adenocarcinoma organoids remains unknown. Here, we examined the ability of PDOs to predict clinical responses to targeted therapies in individual patients and to identify effective anticancer therapies for novel molecular targets.
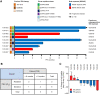
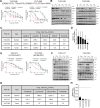
Experimental design: Eighty-four organoids were established from patients with advanced lung adenocarcinoma. Formalin-fixed, paraffin-embedded tumor specimens from corresponding patients were analyzed by whole-exome sequencing (n = 12). Organoids were analyzed by whole-exome sequencing (n = 61) and RNA sequencing (n = 55). Responses to mono or combination targeted therapies were examined in organoids and organoid-derived xenografts.
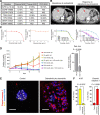
Results: PDOs largely retained somatic alterations including driver mutations of matching patient tumors. PDOs were able to recapitulate progression-free survival and objective responses of patients with non-small cell lung cancer receiving clinically approved tyrosine kinase inhibitors. PDOs recapitulated activity of therapeutic strategies under clinical investigation. YUO-071 harboring an EGFR exon 19 deletion and a BRAF G464A mutation and the matching patient responded to dabrafenib/trametinib combination therapy. YUO-004 and YUO-050 harboring an EGFR L747P mutation was sensitive to afatinib, consistent with the response in the matching patient of YUO-050. Furthermore, we utilized organoids to identify effective therapies for novel molecular targets by demonstrating the efficacy of poziotinib against ERBB2 exon 20 insertions and pralsetinib against RET fusions.
Conclusions: We demonstrated translational relevance of PDOs in advanced lung adenocarcinoma. PDOs are an important diagnostic tool, which can assist clinical decision making and accelerate development of therapeutic strategies.
©2021 The Authors; Published by the American Association for Cancer Research.
Figures



References
-
- Yang C-Y, Yang JC-H, Yang P-C. Precision management of advanced non–small cell lung cancer. Annu Rev Med 2020;71:117–36. - PubMed
-
- National Comprehensive Cancer Network. Non-small cell lung cancer (version 8. 2020). Available from: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf.
-
- Lim SM, Kim HR, Lee J-S, Lee KH, Lee Y-G, Min YJ, et al. . Open-label, multicenter, phase II study of ceritinib in patients with non–small-cell lung cancer harboring ROS1 rearrangement. J Clin Oncol 2017;35:2613–8. - PubMed
-
- Mok TS, Wu Y-L, Thongprasert S, Yang C-H, Chu D-T, Saijo N, et al. . Gefitinib or carboplatin–paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947–57. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

