Polycomb-dependent differential chromatin compartmentalization determines gene coregulation in Arabidopsis
- PMID: 34083408
- PMCID: PMC8256866
- DOI: 10.1101/gr.273771.120
Polycomb-dependent differential chromatin compartmentalization determines gene coregulation in Arabidopsis
Abstract
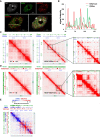
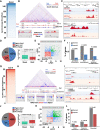
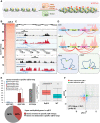
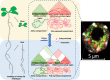
In animals, distant H3K27me3-marked Polycomb targets can establish physical interactions forming repressive chromatin hubs. In plants, growing evidence suggests that H3K27me3 acts directly or indirectly to regulate chromatin interactions, although how this histone modification modulates 3D chromatin architecture remains elusive. To decipher the impact of the dynamic deposition of H3K27me3 on the Arabidopsis thaliana nuclear interactome, we combined genetics, transcriptomics, and several 3D epigenomic approaches. By analyzing mutants defective for histone H3K27 methylation or demethylation, we uncovered the crucial role of this chromatin mark in short- and previously unnoticed long-range chromatin loop formation. We found that a reduction in H3K27me3 levels led to a decrease in the interactions within Polycomb-associated repressive domains. Regions with lower H3K27me3 levels in the H3K27 methyltransferase clf mutant established new interactions with regions marked with H3K9ac, a histone modification associated with active transcription, indicating that a reduction in H3K27me3 levels induces a global reconfiguration of chromatin architecture. Altogether, our results reveal that the 3D genome organization is tightly linked to reversible histone modifications that govern chromatin interactions. Consequently, nuclear organization dynamics shapes the transcriptional reprogramming during plant development and places H3K27me3 as a key feature in the coregulation of distant genes.
© 2021 Huang et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
