Reciprocal repulsions instruct the precise assembly of parallel hippocampal networks
- PMID: 34083484
- PMCID: PMC8830376
- DOI: 10.1126/science.abg1774
Reciprocal repulsions instruct the precise assembly of parallel hippocampal networks
Abstract
Mammalian medial and lateral hippocampal networks preferentially process spatial- and object-related information, respectively. However, the mechanisms underlying the assembly of such parallel networks during development remain largely unknown. Our study shows that, in mice, complementary expression of cell surface molecules teneurin-3 (Ten3) and latrophilin-2 (Lphn2) in the medial and lateral hippocampal networks, respectively, guides the precise assembly of CA1-to-subiculum connections in both networks. In the medial network, Ten3-expressing (Ten3+) CA1 axons are repelled by target-derived Lphn2, revealing that Lphn2- and Ten3-mediated heterophilic repulsion and Ten3-mediated homophilic attraction cooperate to control precise target selection of CA1 axons. In the lateral network, Lphn2-expressing (Lphn2+) CA1 axons are confined to Lphn2+ targets via repulsion from Ten3+ targets. Our findings demonstrate that assembly of parallel hippocampal networks follows a "Ten3→Ten3, Lphn2→Lphn2" rule instructed by reciprocal repulsions.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures


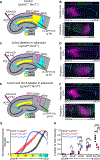
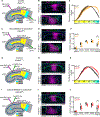
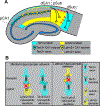
Comment in
-
Attraction and repulsion cooperate during brain-circuit wiring.Nature. 2021 Jun;594(7863):341-343. doi: 10.1038/d41586-021-01502-0. Nature. 2021. PMID: 34089037 No abstract available.
References
-
- O’Keefe J, Dostrovsky J, Brain Res. 34, 171–175 (1971). - PubMed
-
- Scoville WB, Milner B, Neuropsychiatry Clin J. Neurosci. 12, 103–113 (2000). - PubMed
-
- Squire LR, Stark CEL, Clark RE, Annu. Rev. Neurosci 27, 279–306 (2004). - PubMed
-
- Hafting T, Fyhn M, Molden S, Moser M-B, Moser EI, Nature 436, 801–806 (2005). - PubMed
-
- Igarashi KM, Ito HT, Moser EI, Moser M-B, FEBS Lett. 588, 2470–2476 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

