STAT1 potentiates oxidative stress revealing a targetable vulnerability that increases phenformin efficacy in breast cancer
- PMID: 34083537
- PMCID: PMC8175605
- DOI: 10.1038/s41467-021-23396-2
STAT1 potentiates oxidative stress revealing a targetable vulnerability that increases phenformin efficacy in breast cancer
Abstract
Bioenergetic perturbations driving neoplastic growth increase the production of reactive oxygen species (ROS), requiring a compensatory increase in ROS scavengers to limit oxidative stress. Intervention strategies that simultaneously induce energetic and oxidative stress therefore have therapeutic potential. Phenformin is a mitochondrial complex I inhibitor that induces bioenergetic stress. We now demonstrate that inflammatory mediators, including IFNγ and polyIC, potentiate the cytotoxicity of phenformin by inducing a parallel increase in oxidative stress through STAT1-dependent mechanisms. Indeed, STAT1 signaling downregulates NQO1, a key ROS scavenger, in many breast cancer models. Moreover, genetic ablation or pharmacological inhibition of NQO1 using β-lapachone (an NQO1 bioactivatable drug) increases oxidative stress to selectively sensitize breast cancer models, including patient derived xenografts of HER2+ and triple negative disease, to the tumoricidal effects of phenformin. We provide evidence that therapies targeting ROS scavengers increase the anti-neoplastic efficacy of mitochondrial complex I inhibitors in breast cancer.
Conflict of interest statement
The authors declare no competing interests.
Figures
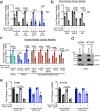

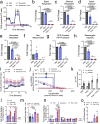

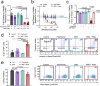


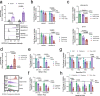

References
-
- Beca F, Polyak K. Intratumor heterogeneity in breast cancer. Adv. Exp. Med. Biol. 2016;882:169–189. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

