Automatic and unbiased segmentation and quantification of myofibers in skeletal muscle
- PMID: 34083673
- PMCID: PMC8175575
- DOI: 10.1038/s41598-021-91191-6
Automatic and unbiased segmentation and quantification of myofibers in skeletal muscle
Abstract
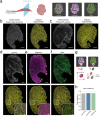
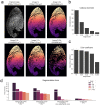
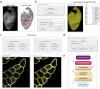
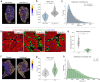
Skeletal muscle has the remarkable ability to regenerate. However, with age and disease muscle strength and function decline. Myofiber size, which is affected by injury and disease, is a critical measurement to assess muscle health. Here, we test and apply Cellpose, a recently developed deep learning algorithm, to automatically segment myofibers within murine skeletal muscle. We first show that tissue fixation is necessary to preserve cellular structures such as primary cilia, small cellular antennae, and adipocyte lipid droplets. However, fixation generates heterogeneous myofiber labeling, which impedes intensity-based segmentation. We demonstrate that Cellpose efficiently delineates thousands of individual myofibers outlined by a variety of markers, even within fixed tissue with highly uneven myofiber staining. We created a novel ImageJ plugin (LabelsToRois) that allows processing of multiple Cellpose segmentation images in batch. The plugin also contains a semi-automatic erosion function to correct for the area bias introduced by the different stainings, thereby identifying myofibers as accurately as human experts. We successfully applied our segmentation pipeline to uncover myofiber regeneration differences between two different muscle injury models, cardiotoxin and glycerol. Thus, Cellpose combined with LabelsToRois allows for fast, unbiased, and reproducible myofiber quantification for a variety of staining and fixation conditions.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

