An in vivo selection system with tightly regulated gene expression enables directed evolution of highly efficient enzymes
- PMID: 34083677
- PMCID: PMC8175713
- DOI: 10.1038/s41598-021-91204-4
An in vivo selection system with tightly regulated gene expression enables directed evolution of highly efficient enzymes
Abstract
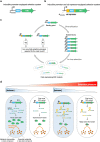
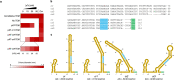
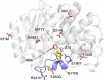
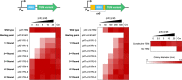
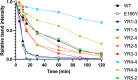
In vivo selection systems are powerful tools for directed evolution of enzymes. The selection pressure of the systems can be tuned by regulating the expression levels of the catalysts. In this work, we engineered a selection system for laboratory evolution of highly active enzymes by incorporating a translationally suppressing cis repressor as well as an inducible promoter to impart stringent and tunable selection pressure. We demonstrated the utility of our selection system by performing directed evolution experiments using TEM β-lactamase as the model enzyme. Five evolutionary rounds afforded a highly active variant exhibiting 440-fold improvement in catalytic efficiency. We also showed that, without the cis repressor, the selection system cannot provide sufficient selection pressure required for evolving highly efficient TEM β-lactamase. The selection system should be applicable for the exploration of catalytic perfection of a wide range of enzymes.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Directed evolution of a protein container.Science. 2011 Feb 4;331(6017):589-92. doi: 10.1126/science.1199081. Science. 2011. PMID: 21292977
-
Acquisition of novel catalytic activity by the M1 RNA ribozyme: the cost of molecular adaptation.J Mol Biol. 1999 Oct 1;292(4):931-44. doi: 10.1006/jmbi.1999.3098. J Mol Biol. 1999. PMID: 10525416
-
A simple selection strategy for evolving highly efficient enzymes.Nat Biotechnol. 2007 Oct;25(10):1145-7. doi: 10.1038/nbt1341. Epub 2007 Sep 16. Nat Biotechnol. 2007. PMID: 17873865
-
Genetic selection as a tool in mechanistic enzymology and protein design.Ernst Schering Res Found Workshop. 2000;(32):253-68. doi: 10.1007/978-3-662-04042-3_9. Ernst Schering Res Found Workshop. 2000. PMID: 11077612 Review. No abstract available.
-
FNR and its role in oxygen-regulated gene expression in Escherichia coli.FEMS Microbiol Rev. 1990 Aug;6(4):399-428. doi: 10.1111/j.1574-6968.1990.tb04109.x. FEMS Microbiol Rev. 1990. PMID: 2248796 Review.
Cited by
-
A growth selection system for sucrose synthases (SuSy): design and test.AMB Express. 2024 Jun 12;14(1):70. doi: 10.1186/s13568-024-01727-y. AMB Express. 2024. PMID: 38865019 Free PMC article.
-
A growth selection system for the directed evolution of amine-forming or converting enzymes.Nat Commun. 2022 Dec 3;13(1):7458. doi: 10.1038/s41467-022-35228-y. Nat Commun. 2022. PMID: 36460668 Free PMC article.
-
Directed Evolution of 4-Hydroxyphenylpyruvate Biosensors Based on a Dual Selection System.Int J Mol Sci. 2024 Jan 26;25(3):1533. doi: 10.3390/ijms25031533. Int J Mol Sci. 2024. PMID: 38338812 Free PMC article.
-
Growth-Coupled Evolutionary Pressure Improving Epimerases for D-Allulose Biosynthesis Using a Biosensor-Assisted In Vivo Selection Platform.Adv Sci (Weinh). 2024 Apr;11(14):e2306478. doi: 10.1002/advs.202306478. Epub 2024 Feb 2. Adv Sci (Weinh). 2024. PMID: 38308132 Free PMC article.
-
A growth-based screening strategy for engineering the catalytic activity of an oxygen-sensitive formate dehydrogenase.Appl Environ Microbiol. 2024 Sep 18;90(9):e0147224. doi: 10.1128/aem.01472-24. Epub 2024 Aug 28. Appl Environ Microbiol. 2024. PMID: 39194220 Free PMC article.
References
-
- Kast P, Hilvert D. Genetic selection strategies for generating and characterizing catalysts. Pure Appl. Chem. 1996;68:2017–2024. doi: 10.1351/pac199668112017. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

