BET inhibitor suppresses migration of human hepatocellular carcinoma by inhibiting SMARCA4
- PMID: 34083693
- PMCID: PMC8175750
- DOI: 10.1038/s41598-021-91284-2
BET inhibitor suppresses migration of human hepatocellular carcinoma by inhibiting SMARCA4
Abstract
Hepatocellular carcinoma (HCC) is one of the most prevalent and poorly responsive cancers worldwide. Bromodomain and extraterminal (BET) inhibitors, such as JQ1 and OTX-015, inhibit BET protein binding to acetylated residues in histones. However, the physiological mechanisms and regulatory processes of BET inhibition in HCC remain unclear. To explore BET inhibitors' potential role in the molecular mechanisms underlying their anticancer effects in HCC, we analyzed BET inhibitor-treated HCC cells' gene expression profiles with RNA-seq and bioinformatics analysis. BET inhibitor treatment significantly downregulated genes related to bromodomain-containing proteins 4 (BRD4), such as ACSL5, SLC38A5, and ICAM2. Importantly, some cell migration-related genes, including AOC3, CCR6, SSTR5, and SCL7A11, were significantly downregulated. Additionally, bioinformatics analysis using Ingenuity Knowledge Base Ingenuity Pathway Analysis (IPA) revealed that SMARCA4 regulated migration response molecules. Furthermore, knockdown of SMARCA4 gene expression by siRNA treatment significantly reduced cell migration and the expression of migration-related genes. In summary, our results indicated that BET inhibitor treatment in HCC cell lines reduces cell migration through the downregulation of SMARCA4.
Conflict of interest statement
The authors declare no competing interests.
Figures
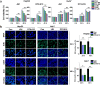


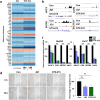

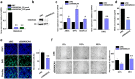

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

