A conserved rhizobial peptidase that interacts with host-derived symbiotic peptides
- PMID: 34083727
- PMCID: PMC8175422
- DOI: 10.1038/s41598-021-91394-x
A conserved rhizobial peptidase that interacts with host-derived symbiotic peptides
Abstract
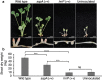
In the Medicago truncatula-Sinorhizobium meliloti symbiosis, chemical signaling initiates rhizobial infection of root nodule tissue, where a large portion of the bacteria are endocytosed into root nodule cells to function in nitrogen-fixing organelles. These intracellular bacteria are subjected to an arsenal of plant-derived nodule-specific cysteine-rich (NCR) peptides, which induce the physiological changes that accompany nitrogen fixation. NCR peptides drive these intracellular bacteria toward terminal differentiation. The bacterial peptidase HrrP was previously shown to degrade host-derived NCR peptides and give the bacterial symbionts greater fitness at the expense of host fitness. The hrrP gene is found in roughly 10% of Sinorhizobium isolates, as it is carried on an accessory plasmid. The objective of the present study is to identify peptidase genes in the core genome of S. meliloti that modulate symbiotic outcome in a manner similar to the accessory hrrP gene. In an overexpression screen of annotated peptidase genes, we identified one such symbiosis-associated peptidase (sap) gene, sapA (SMc00451). When overexpressed, sapA leads to a significant decrease in plant fitness. Its promoter is active in root nodules, with only weak expression evident under free-living conditions. The SapA enzyme can degrade a broad range of NCR peptides in vitro.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

