Clock-in, clock-out: circadian timekeeping between tissues
- PMID: 34083887
- PMCID: PMC8171283
- DOI: 10.1042/bio04202007
Clock-in, clock-out: circadian timekeeping between tissues
Abstract
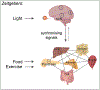
Life evolved in the presence of alternating periods of light and dark that accompany the daily rotation of the Earth on its axis. This offered an advantage for organisms able to regulate their physiology to anticipate these daily cycles. In each light-sensitive organism studied, spanning single-celled bacteria to complex mammals, there exist timekeeping mechanisms able to control physiology over the course of 24 hours. Endowed with internal timekeeping, organisms can put their previously stored energy to the most efficient use, selectively ramping up biological processes at specific times of day or night according to when they'll be needed. Humans have evolved to be more active during the day (diurnal), likely due to the increased opportunities for foraging or hunting in our evolutionary past, and this daily activity is accompanied by an upregulation of genes involved in metabolism to increase the energy available for such behaviours. Remarkably, this happens without conscious thought, due to a complex organism-wide signalling apparatus known as the circadian clock network, that conveys time information between cells and tissues.
Figures



References
-
- Crosby P, Hamnett R, Putker M, Hoyle NP, Reed M, Karam CJ, Maywood ES, Stangherlin A, Chesham JE, Hayter EA, Rosenbrier-Ribeiro L, Newham P, Clevers H, Bechtold DA, O'Neill JS. Insulin/IGF-1 Drives PERIOD Synthesis to Entrain Circadian Rhythms with Feeding Time. Cell. 2019. May 2;177(4):896–909.e20. doi: 10.1016/j.cell.2019.02.017. Epub 2019 Apr 25. - DOI - PMC - PubMed
-
- Dyar KA, Lutter D, Artati A, Ceglia NJ, Liu Y, Armenta D, Jastroch M, Schneider S, de Mateo S, Cervantes M, Abbondante S, Tognini P, Orozco-Solis R, Kinouchi K, Wang C, Swerdloff R, Nadeef S, Masri S, Magistretti P, Orlando V, Borrelli E, Uhlenhaut NH, Baldi P, Adamski J, Tschöp MH, Eckel-Mahan K, Sassone-Corsi P. Atlas of Circadian Metabolism Reveals System-wide Coordination and Communication between Clocks. Cell. 2018. September 6;174(6):1571–1585.e11. doi: 10.1016/j.cell.2018.08.042. - DOI - PMC - PubMed
-
- Perrin L, Loizides-Mangold U, Skarupelova S, Pulimeno P, Chanon S, Robert M, Bouzakri K, Modoux C, Roux-Lombard P, Vidal H, Lefai E, Dibner C. Human skeletal myotubes display a cell-autonomous circadian clock implicated in basal myokine secretion. Mol Metab. 2015. August 6;4(11):834–45. doi: 10.1016/j.molmet.2015.07.009. eCollection 2015 November. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
