The Chromatin Accessibility Landscape of Peripheral Blood Mononuclear Cells in Patients With Systemic Lupus Erythematosus at Single-Cell Resolution
- PMID: 34084162
- PMCID: PMC8168536
- DOI: 10.3389/fimmu.2021.641886
The Chromatin Accessibility Landscape of Peripheral Blood Mononuclear Cells in Patients With Systemic Lupus Erythematosus at Single-Cell Resolution
Abstract
Objective: Systemic lupus erythematosus (SLE) is a complex autoimmune disease, and various immune cells are involved in the initiation, progression, and regulation of SLE. Our goal was to reveal the chromatin accessibility landscape of peripheral blood mononuclear cells (PBMCs) in SLE patients at single-cell resolution and identify the transcription factors (TFs) that may drive abnormal immune responses.
Methods: The assay for transposase accessible chromatin in single-cell sequencing (scATAC-seq) method was applied to map the landscape of active regulatory DNA in immune cells from SLE patients at single-cell resolution, followed by clustering, peak annotation and motif analysis of PBMCs in SLE.
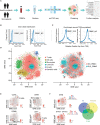
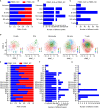
Results: Peripheral blood mononuclear cells were robustly clustered based on their types without using antibodies. We identified twenty patterns of TF activation that drive abnormal immune responses in SLE patients. Then, we observed ten genes that were highly associated with SLE pathogenesis by altering T cell activity. Finally, we found 12 key TFs regulating the above six genes (CD83, ELF4, ITPKB, RAB27A, RUNX3, and ZMIZ1) that may be related to SLE disease pathogenesis and were significantly enriched in SLE patients (p <0.05, FC >2). With qPCR experiments on CD83, ELF4, RUNX3, and ZMIZ1 in B cells, we observed a significant difference in the expression of genes (ELF4, RUNX3, and ZMIZ1), which were regulated by seven TFs (EWSR1-FLI1, MAF, MAFA, NFIB, NR2C2 (var. 2), TBX4, and TBX5). Meanwhile, the seven TFs showed highly accessible binding sites in SLE patients.
Conclusions: These results confirm the importance of using single-cell sequencing to uncover the real features of immune cells in SLE patients, reveal key TFs in SLE-PBMCs, and provide foundational insights relevant for epigenetic therapy.
Keywords: marker; peripheral blood mononuclear cells; single-cell chromatin accessible assay; systemic lupus erythematosus; transcription factor.
Copyright © 2021 Yu, Hong, Wu, Zheng, Zeng, Dai, Yin, Liu, Tang and Dai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

