Apoptotic vesicles restore liver macrophage homeostasis to counteract type 2 diabetes
- PMID: 34084287
- PMCID: PMC8144839
- DOI: 10.1002/jev2.12109
Apoptotic vesicles restore liver macrophage homeostasis to counteract type 2 diabetes
Abstract
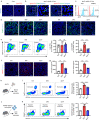
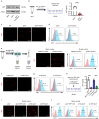
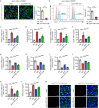
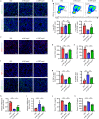
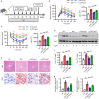
Apoptosis is a naturally occurring process generating plenty of apoptotic vesicles (apoVs), but the feature, fate and function of apoVs remain largely unknown. Notably, as an appealing source for cell therapy, mesenchymal stem cells (MSCs) undergo necessary apoptosis and release apoVs during therapeutic application. In this study, we characterized and used MSC-derived apoVs to treat type 2 diabetes (T2D) mice, and we found that apoVs were efferocytosed by macrophages and functionally modulated liver macrophage homeostasis to counteract T2D. We showed that apoVs can induce macrophage reprogramming at the transcription level in an efferocytosis-dependent manner, leading to inhibition of macrophage accumulation and transformation of macrophages towards an anti-inflammation phenotype in T2D liver. At the molecular level, we discovered that calreticulin (CRT) was exposed on the surface of apoVs to act as a critical 'eat-me' signal mediating apoV efferocytosis and macrophage regulatory effects. Importantly, we demonstrated that CRT-mediated efferocytosis of MSC-derived apoVs contributes to T2D therapy with alleviation of T2D phenotypes including glucose intolerance and insulin resistance. These findings uncover that functional efferocytosis of apoVs restores liver macrophage homeostasis and ameliorates T2D.
Keywords: apoptotic vesicles; calreticulin; efferocytosis; macrophages; mesenchymal stem cells; type 2 diabetes.
© 2021 The Authors. Journal of Extracellular Vesicles published by Wiley Periodicals, LLC on behalf of the International Society for Extracellular Vesicles.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Baeck, C. , Wehr, A. , Karlmark, K. R. , Heymann, F. , Vucur, M. , Gassler, N. , Huss, S. , Klussmann, S. , Eulberg, D. , Luedde, T. , Trautwein, C. , & Tacke, F. (2012). Pharmacological inhibition of the chemokine CCL2 (MCP‐1) diminishes liver macrophage infiltration and steatohepatitis in chronic hepatic injury. Gut, 61(3), 416–426. - PubMed
-
- Barnes, E. , Ward, S M. , Kasprowicz, V O. , Dusheiko, G. , Klenerman, P. , & Lucas, M. (2004). Ultra‐sensitive class I tetramer analysis reveals previously undetectable populations of antiviral CD8+ T cells. European Journal of Immunology, 34(6), 1570–1577. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

