Large scale relative protein ligand binding affinities using non-equilibrium alchemy
- PMID: 34084371
- PMCID: PMC8145179
- DOI: 10.1039/c9sc03754c
Large scale relative protein ligand binding affinities using non-equilibrium alchemy
Abstract
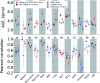
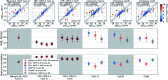
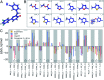
Ligand binding affinity calculations based on molecular dynamics (MD) simulations and non-physical (alchemical) thermodynamic cycles have shown great promise for structure-based drug design. However, their broad uptake and impact is held back by the notoriously complex setup of the calculations. Only a few tools other than the free energy perturbation approach by Schrödinger Inc. (referred to as FEP+) currently enable end-to-end application. Here, we present for the first time an approach based on the open-source software pmx that allows to easily set up and run alchemical calculations for diverse sets of small molecules using the GROMACS MD engine. The method relies on theoretically rigorous non-equilibrium thermodynamic integration (TI) foundations, and its flexibility allows calculations with multiple force fields. In this study, results from the Amber and Charmm force fields were combined to yield a consensus outcome performing on par with the commercial FEP+ approach. A large dataset of 482 perturbations from 13 different protein-ligand datasets led to an average unsigned error (AUE) of 3.64 ± 0.14 kJ mol-1, equivalent to Schrödinger's FEP+ AUE of 3.66 ± 0.14 kJ mol-1. For the first time, a setup is presented for overall high precision and high accuracy relative protein-ligand alchemical free energy calculations based on open-source software.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures





References
-
- Wang L. Wu Y. Deng Y. Kim B. Pierce L. Krilov G. Lupyan D. Robinson S. Dahlgren M. K. Greenwood J. Romero D. L. Masse C. Knight J. L. Steinbrecher T. Beuming T. Damm W. Harder E. Sherman W. Brewer M. Wester R. Murcko M. Frye L. Farid R. Lin T. Mobley D. L. Jorgensen W. L. Berne B. J. Friesner R. A. Abel R. J. Am. Chem. Soc. 2015;137:2695–2703. doi: 10.1021/ja512751q. - DOI - PubMed
LinkOut - more resources
Full Text Sources

