Mitochondrial autophagy and cell survival is regulated by the circadian Clock gene in cardiac myocytes during ischemic stress
- PMID: 34085589
- PMCID: PMC8632283
- DOI: 10.1080/15548627.2021.1938913
Mitochondrial autophagy and cell survival is regulated by the circadian Clock gene in cardiac myocytes during ischemic stress
Abstract
Cardiac function is highly reliant on mitochondrial oxidative metabolism and quality control. The circadian Clock gene is critically linked to vital physiological processes including mitochondrial fission, fusion and bioenergetics; however, little is known of how the Clock gene regulates these vital processes in the heart. Herein, we identified a putative circadian CLOCK-mitochondrial interactome that gates an adaptive survival response during myocardial ischemia. We show by transcriptome and gene ontology mapping in CLOCK Δ19/Δ19 mouse that Clock transcriptionally coordinates the efficient removal of damaged mitochondria during myocardial ischemia by directly controlling transcription of genes required for mitochondrial fission, fusion and macroautophagy/autophagy. Loss of Clock gene activity impaired mitochondrial turnover resulting in the accumulation of damaged reactive oxygen species (ROS)-producing mitochondria from impaired mitophagy. This coincided with ultrastructural defects to mitochondria and impaired cardiac function. Interestingly, wild type CLOCK but not mutations of CLOCK defective for E-Box binding or interaction with its cognate partner ARNTL/BMAL-1 suppressed mitochondrial damage and cell death during acute hypoxia. Interestingly, the autophagy defect and accumulation of damaged mitochondria in CLOCK-deficient cardiac myocytes were abrogated by restoring autophagy/mitophagy. Inhibition of autophagy by ATG7 knockdown abrogated the cytoprotective effects of CLOCK. Collectively, our results demonstrate that CLOCK regulates an adaptive stress response critical for cell survival by transcriptionally coordinating mitochondrial quality control mechanisms in cardiac myocytes. Interdictions that restore CLOCK activity may prove beneficial in reducing cardiac injury in individuals with disrupted circadian CLOCK.Abbreviations: ARNTL/BMAL1: aryl hydrocarbon receptor nuclear translocator-like; ATG14: autophagy related 14; ATG7: autophagy related 7; ATP: adenosine triphosphate; BCA: bovine serum albumin; BECN1: beclin 1, autophagy related; bHLH: basic helix- loop-helix; CLOCK: circadian locomotor output cycles kaput; CMV: cytomegalovirus; COQ5: coenzyme Q5 methyltransferase; CQ: chloroquine; CRY1: cryptochrome 1 (photolyase-like); DNM1L/DRP1: dynamin 1-like; EF: ejection fraction; EM: electron microscopy; FS: fractional shortening; GFP: green fluorescent protein; HPX: hypoxia; i.p.: intraperitoneal; I-R: ischemia-reperfusion; LAD: left anterior descending; LVIDd: left ventricular internal diameter diastolic; LVIDs: left ventricular internal diameter systolic; MAP1LC3/LC3: microtubule-associated protein 1 light chain 3; MFN2: mitofusin 2; MI: myocardial infarction; mPTP: mitochondrial permeability transition pore; NDUFA4: Ndufa4, mitochondrial complex associated; NDUFA8: NADH: ubiquinone oxidoreductase subunit A8; NMX: normoxia; OCR: oxygen consumption rate; OPA1: OPA1, mitochondrial dynamin like GTPase; OXPHOS: oxidative phosphorylation; PBS: phosphate-buffered saline; PER1: period circadian clock 1; PPARGC1A/PGC-1α: peroxisome proliferative activated receptor, gamma, coactivator 1 alpha; qPCR: quantitative real-time PCR; RAB7A: RAB7, member RAS oncogene family; ROS: reactive oxygen species; RT: room temperature; shRNA: short hairpin RNA; siRNA: small interfering RNA; TFAM: transcription factor A, mitochondrial; TFEB: transcription factor EB; TMRM: tetra-methylrhodamine methyl ester perchlorate; WT: wild -type; ZT: zeitgeber time.
Keywords: Autophagy; clock; metabolism; mitochondrion; myocardial infarction.
Conflict of interest statement
The authors declare no competing interests.
Figures
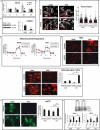
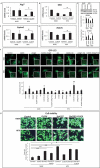
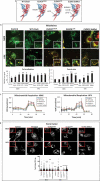
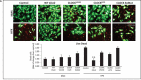
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
