Contribution of adhesion proteins to Aggregatibacter actinomycetemcomitans biofilm formation
- PMID: 34085776
- PMCID: PMC8349852
- DOI: 10.1111/omi.12346
Contribution of adhesion proteins to Aggregatibacter actinomycetemcomitans biofilm formation
Abstract
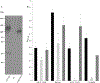
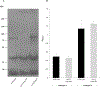
Aggregatibacter actinomycetemcomitans is a Gram-negative bacterium associated with periodontal disease and multiple disseminated extra-oral infections. Colonization of these distinct physiological niches is contingent on the expression of specific surface proteins during the initiation of developing biofilms. In this investigation, we studied fimbriae and three well-characterized nonfimbrial surface proteins (EmaA, Aae, and ApiA/Omp100) for their contribution to biofilm formation. Mutations of these proteins in multiple strains covering four different serotypes demonstrated variance in biofilm development that was strain dependent but independent of serotype. In a fimbriated background, only inactivation of emaA impacted biofilm mass. In contrast, inactivation of emaA and/or aae affected biofilm formation in nonfimbriated A. actinomycetemcomitans strains, whereas inactivation of apiA/omp100 had little effect on biofilm formation. When these genes were expressed individually in Escherichia coli, all transformed strains demonstrated an increase in biofilm mass compared to the parent strain. The strain expressing emaA generated the greatest mass of biofilm, whereas the strains expressing either aae or apiA/omp100 were greatly reduced and similar in mass. These data suggest a redundancy in function of these nonfimbrial adhesins, which is dependent on the genetic background of the strain investigated.
Keywords: adhesins; autotransporter proteins; biofilm; periodontitis.
© 2021 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Figures



References
-
- Alugupalli KR, Kalfas S, & Forsgren A. (1996). Laminin binding to a heat-modifiable outer membrane protein of Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol, 11(5), 326–331. - PubMed
-
- Asakawa R, Komatsuzawa H, Kawai T, et al. (2003). Outer membrane protein 100, a versatile virulence factor of Actinobacillus actinomycetemcomitans. Mol Microbiol, 50(4), 1125–1139. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

