In lysate RNA digestion provides insights into the angiogenin's specificity towards transfer RNAs
- PMID: 34085908
- PMCID: PMC8632075
- DOI: 10.1080/15476286.2021.1930758
In lysate RNA digestion provides insights into the angiogenin's specificity towards transfer RNAs
Abstract
Under adverse conditions, tRNAs are processed into fragments called tRNA-derived stress-induced RNAs (tiRNAs) by stress-responsive ribonucleases (RNases) such as angiogenin (ANG). Recent studies have reported several biological functions of synthetic tiRNAs lacking post-transcriptional modifications found on endogenous tiRNAs. Here we describe a simple and reproducible method to efficiently isolate ANG-cleaved tiRNAs from endogenous tRNAs. Using this in vitro method, more than 50% of mature tRNAs are cleaved into tiRNAs which can be enriched using complementary oligonucleotides. Using this method, the yield of isolated endogenous 5'-tiRNAGly-GCC was increased about fivefold compared to when tiRNAs were obtained by cellular treatment of ANG. Although the non-specific ribonuclease activity of ANG is much lower than that of RNase A, we show that ANG cleaves physiologically folded tRNAs as efficiently as bovine RNase A. These results suggest that ANG is highly specialized to cleave physiologically folded tRNAs. Our method will greatly facilitate the analysis of endogenous tiRNAs to elucidate the physiological functions of ANG.
Keywords: stress; tRNA; tRNA-derived fragments; tiRNAs.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
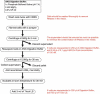
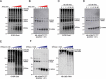
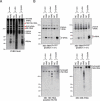


References
-
- Sun C, Fu Z, Wang S, et al. Roles of tRNA-derived fragments in human cancers. Cancer Lett. 2018;414:16–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
