Predicting RNA Scaffolds with a Hybrid Method of Vfold3D and VfoldLA
- PMID: 34086269
- PMCID: PMC9728534
- DOI: 10.1007/978-1-0716-1499-0_1
Predicting RNA Scaffolds with a Hybrid Method of Vfold3D and VfoldLA
Abstract
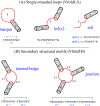
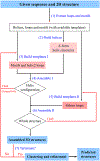
The ever-increasing discoveries of noncoding RNA functions draw a strong demand for RNA structure determination from the sequence. In recently years, computational studies for RNA structures, at both the two-dimensional and the three-dimensional levels, led to several highly promising new developments. In this chapter, we describe a hybrid method, which combines the motif template-based Vfold3D model and the loop template-based VfoldLA model, to predict RNA 3D structures. The main emphasis is placed on the definition of motifs and loops, the treatment of no-template motifs, and the 3D structure assembly from templates of motifs and loops. For illustration, we use the ZIKV xrRNA1 as an example to show the template-based prediction of RNA 3D structures from the 2D structure. The web server for the hybrid model is freely accessible at http://rna.physics.missouri.edu/vfold3D2 .
Keywords: Secondary structural motifs; Single-stranded loops; Structure prediction; Template-assembly.
Figures

Similar articles
-
VfoldLA: A web server for loop assembly-based prediction of putative 3D RNA structures.J Struct Biol. 2019 Sep 1;207(3):235-240. doi: 10.1016/j.jsb.2019.06.002. Epub 2019 Jun 4. J Struct Biol. 2019. PMID: 31173857 Free PMC article.
-
A Method to Predict the 3D Structure of an RNA Scaffold.Methods Mol Biol. 2015;1316:1-11. doi: 10.1007/978-1-4939-2730-2_1. Methods Mol Biol. 2015. PMID: 25967048 Free PMC article.
-
Hierarchical Assembly of RNA Three-Dimensional Structures Based on Loop Templates.J Phys Chem B. 2018 May 31;122(21):5327-5335. doi: 10.1021/acs.jpcb.7b10102. Epub 2018 Jan 8. J Phys Chem B. 2018. PMID: 29258305 Free PMC article.
-
Functional RNA during Zika virus infection.Virus Res. 2018 Aug 2;254:41-53. doi: 10.1016/j.virusres.2017.08.015. Epub 2017 Aug 31. Virus Res. 2018. PMID: 28864425 Review.
-
Genomic characterization and phylogenetic analysis of Zika virus circulating in the Americas.Infect Genet Evol. 2016 Sep;43:43-9. doi: 10.1016/j.meegid.2016.05.004. Epub 2016 May 6. Infect Genet Evol. 2016. PMID: 27156653 Review.
Cited by
-
RNA 3D Structure Prediction: Progress and Perspective.Molecules. 2023 Jul 20;28(14):5532. doi: 10.3390/molecules28145532. Molecules. 2023. PMID: 37513407 Free PMC article. Review.
-
Structural Prediction of Coronavirus s2m Kissing Complexes and Extended Duplexes.ACS Phys Chem Au. 2025 Jun 5;5(4):410-424. doi: 10.1021/acsphyschemau.5c00031. eCollection 2025 Jul 23. ACS Phys Chem Au. 2025. PMID: 40727223 Free PMC article.
-
Systematic benchmarking of deep-learning methods for tertiary RNA structure prediction.PLoS Comput Biol. 2024 Dec 30;20(12):e1012715. doi: 10.1371/journal.pcbi.1012715. eCollection 2024 Dec. PLoS Comput Biol. 2024. PMID: 39775239 Free PMC article.
References
-
- Eddy SR (2001) Non-coding RNA genes and the modem RNA world. Nat Rev Genet 2: 919–929 - PubMed
-
- Sharp PA (2009) The centrality of RNA. Cell 136: 577–580 - PubMed
-
- Mortimer SA, Kidwell MA, Doudna JA (2014) Insights into RNA structure and function from genome-wide studies. Nat Rev Genet 15: 469–479 - PubMed
-
- Cech TR, Steitz JA (2014) The noncoding RNA revolution: trashing old rules to forge new ones. Cell 157: 77–94 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

