Characterization of Brassica rapa metallothionein and phytochelatin synthase genes potentially involved in heavy metal detoxification
- PMID: 34086824
- PMCID: PMC8177407
- DOI: 10.1371/journal.pone.0252899
Characterization of Brassica rapa metallothionein and phytochelatin synthase genes potentially involved in heavy metal detoxification
Erratum in
-
Correction: Characterization of Brassica rapa metallothionein and phytochelatin synthase genes potentially involved in heavy metal detoxification.PLoS One. 2022 Jan 27;17(1):e0263427. doi: 10.1371/journal.pone.0263427. eCollection 2022. PLoS One. 2022. PMID: 35085369 Free PMC article.
Abstract
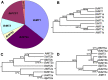
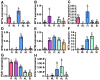
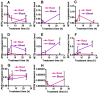
Brassica rapa is an important leafy vegetable that can potentially accumulate high concentrations of cadmium (Cd), posing a risk to human health. The aim of the present study was to identify cadmium detoxifying molecular mechanisms in B. rapa using a functional cloning strategy. A cDNA library constructed from roots of B. rapa plants treated with Cd was transformed into the Cd sensitive yeast mutant strain DTY167 that lacks the yeast cadmium factor (YCF1), and resistant yeast clones were selected on Cd containing media. Two hundred genes potentially conferring cadmium resistance were rescued from the surviving yeast clones and sequenced. Sequencing analysis revealed that genes encoding for metallothionein (MT)1, MT2a, MT2b and MT3, and phytochelatin synthase (PCS)1 and PCS2 accounted for 35.5%, 28.5%, 4%, 11.3%, 18.7% and 2%, respectively of the genes identified. MTs and PCSs expressing DTY167 cells showed resistance to Cd as well as to Zn. PCS1 expressing yeast cells were also more resistant to Pb compared to those expressing MTs or PCS2. RT-PCR results showed that Cd treatment strongly induced the expression levels of MTs in the root and shoot. Furthermore, the different MTs and PCSs exhibited tissue specific expression. The results indicate that MTs and PCS genes potentially play a central role in detoxifying Cd and other toxic metals in B. rapa.
Conflict of interest statement
The authors declare that no conflicting interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

