Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma
- PMID: 34087162
- PMCID: PMC8366750
- DOI: 10.1016/j.ccell.2021.05.002
Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma
Erratum in
-
Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma.Cancer Cell. 2025 Jun 9;43(6):1175-1176. doi: 10.1016/j.ccell.2025.05.007. Epub 2025 May 27. Cancer Cell. 2025. PMID: 40436018 No abstract available.
Abstract
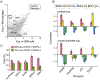
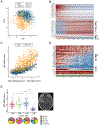
The mesenchymal subtype of glioblastoma is thought to be determined by both cancer cell-intrinsic alterations and extrinsic cellular interactions, but remains poorly understood. Here, we dissect glioblastoma-to-microenvironment interactions by single-cell RNA sequencing analysis of human tumors and model systems, combined with functional experiments. We demonstrate that macrophages induce a transition of glioblastoma cells into mesenchymal-like (MES-like) states. This effect is mediated, both in vitro and in vivo, by macrophage-derived oncostatin M (OSM) that interacts with its receptors (OSMR or LIFR) in complex with GP130 on glioblastoma cells and activates STAT3. We show that MES-like glioblastoma states are also associated with increased expression of a mesenchymal program in macrophages and with increased cytotoxicity of T cells, highlighting extensive alterations of the immune microenvironment with potential therapeutic implications.
Keywords: GBM; OSM; glioblastoma; macrophage; mesenchymal; scRNA-seq; tumor microenvironment.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.L.S. and K.W.W. are equity holders, scientific co-founders, and advisory board members of Immunitas Therapeutics. I.T. is advisory board member of Immunitas Therapeutics. A.R. is a founder and equity holder of Celsius Therapeutics, an equity holder in Immunitas Therapeutics, and until July 31, 2020, was an SAB member for Thermo Fisher Scientific, Syros Pharmaceuticals, Asimov and Neogene Therapeutics. O.R.R. and A.R. are employees of Genentech since October 19, 2020, and August 1, 2020, respectively. K.W.W. serves on the scientific advisory board of TCR2 Therapeutics, T-Scan Therapeutics, SQZ Biotech, and Nextechinvest, and receives sponsored research funding from Novartis. N.D.M. serves as a scientific advisor to Immunitas Therapeutics. X.Z. is a co-founder and consultant for Vizgen, Inc.
Figures




Comment in
-
Macrophage-tumor cell intertwine drives the transition into a mesenchymal-like cellular state of glioblastoma.Cancer Cell. 2021 Jun 14;39(6):743-745. doi: 10.1016/j.ccell.2021.05.003. Epub 2021 Jun 3. Cancer Cell. 2021. PMID: 34087163
References
-
- Butowski N, Colman H, De Groot JF, Omuro AM, Nayak L, Wen PY, Cloughesy TF, Marimuthu A, Haidar S, Perry A, et al. (2016). Orally administered colony stimulating factor 1 receptor inhibitor PLX3397 in recurrent glioblastoma: an Ivy Foundation Early Phase Clinical Trials Consortium phase II study. Neuro-oncology 18, 557–564. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

