Osmolytes and crowders regulate aggregation of the cancer-related L106R mutant of the Axin protein
- PMID: 34087214
- PMCID: PMC8391023
- DOI: 10.1016/j.bpj.2021.05.024
Osmolytes and crowders regulate aggregation of the cancer-related L106R mutant of the Axin protein
Abstract
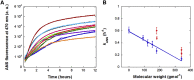
Protein aggregation is involved in a variety of diseases, including neurodegenerative diseases and cancer. The cellular environment is crowded by a plethora of cosolutes comprising small molecules and biomacromolecules at high concentrations, which may influence the aggregation of proteins in vivo. To account for the effect of cosolutes on cancer-related protein aggregation, we studied their effect on the aggregation of the cancer-related L106R mutant of the Axin protein. Axin is a key player in the Wnt signaling pathway, and the L106R mutation in its RGS domain results in a native molten globule that tends to form native-like aggregates. This results in uncontrolled activation of the Wnt signaling pathway, leading to cancer. We monitored the aggregation process of Axin RGS L106R in vitro in the presence of a wide ensemble of cosolutes including polyols, amino acids, betaine, and polyethylene glycol crowders. Except myo-inositol, all polyols decreased RGS L106R aggregation, with carbohydrates exerting the strongest inhibition. Conversely, betaine and polyethylene glycols enhanced aggregation. These results are consistent with the reported effects of osmolytes and crowders on the stability of molten globular proteins and with both amorphous and amyloid aggregation mechanisms. We suggest a model of Axin L106R aggregation in vivo, whereby molecularly small osmolytes keep the protein as a free soluble molecule but the increased crowding of the bound state by macromolecules induces its aggregation at the nanoscale. To our knowledge, this is the first systematic study on the effect of osmolytes and crowders on a process of native-like aggregation involved in pathology, as it sheds light on the contribution of cosolutes to the onset of cancer as a protein misfolding disease and on the relevance of aggregation in the molecular etiology of cancer.
Copyright © 2021 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures









References
-
- Hipp M.S., Kasturi P., Hartl F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019;20:421–435. - PubMed
-
- Chiti F., Dobson C.M. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 2017;86:27–68. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

