Neuropilin-1 assists SARS-CoV-2 infection by stimulating the separation of Spike protein S1 and S2
- PMID: 34087218
- PMCID: PMC8169233
- DOI: 10.1016/j.bpj.2021.05.026
Neuropilin-1 assists SARS-CoV-2 infection by stimulating the separation of Spike protein S1 and S2
Abstract
The cell surface receptor Neuropilin-1 (Nrp1) was recently identified as a host factor for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) entry. The Spike protein of SARS-CoV-2 is cleaved into two segments, the S1 (residues (res.) 1-685) and the S2 (res. 686-1273) domains by furin protease. Nrp1 predominantly binds to the C-terminal RRAR amino acid motif (res. 682-685) of the S1 domain. In this study, we firstly modeled the association of an Nrp1 protein (consisting of domains a2-b1-b2) with the Spike protein. Next, we studied the separation of S2 from the S1 domain, with and without Nrp1 bound, by utilizing molecular dynamics pulling simulations. During the separation, Nrp1 stabilizes the S1 C-terminal region (res. 640-685) and thereby assists the detachment of S2 N-terminal region (res. 686-700). Without Nrp1 bound, S1 tends to become stretched, whereas the bound Nrp1 stimulates an earlier separation of S2 from the S1 domain. The liberated S2 domain is known to mediate the fusion of virus and host membranes; thus, Nrp1 likely increases virus infectivity by facilitating the S1 and S2 separation. We further analyzed the possible topological structure of the SARS-CoV-2 Spike protein when bound with Nrp1 and angiotensin-converting enzyme 2 (ACE2). Understanding of such an Nrp1-assisted viral infection opens the gate for the generation of protein-protein inhibitors, such as antibodies, which could attenuate the infection mechanism and protect certain cells in a future Nrp1-ACE2 targeted combination therapy.
Copyright © 2021 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


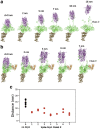

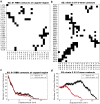
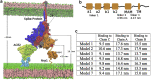
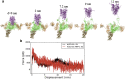
Update of
-
Neuropilin-1 Assists SARS-CoV-2 Infection by Stimulating the Separation of Spike Protein Domains S1 and S2.bioRxiv [Preprint]. 2021 Jan 19:2021.01.06.425627. doi: 10.1101/2021.01.06.425627. bioRxiv. 2021. Update in: Biophys J. 2021 Jul 20;120(14):2828-2837. doi: 10.1016/j.bpj.2021.05.026. PMID: 33442700 Free PMC article. Updated. Preprint.
References
-
- Vankadari N. Structure of furin protease binding to SARS-CoV-2 spike glycoprotein and implications for potential targets and virulence. J. Phys. Chem. Lett. 2020;11:6655–6663. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

